|

Tools
Purchase CGAP Reagents
Related Links
Quick Links:

|
CCAP BAC Clones
Chromosome 14
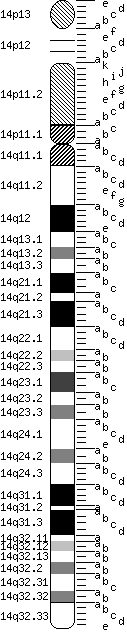
|
|
Map Coordinate
|
BAC id
|
Placement on genome1
|
Placement summary3
|
BAC End2
|
Insert Sequence2
|
STS id2
|
|
|
14q11.1b |
RP11-324B11 |
chr14 19Mb |      |
AZ081795 T7+,
AZ081805 SP6,
AQ539031 SP6 | | RH53565,
WI-18729 |
|
|
14q11.2b |
RP11-332N6 |
chr14 19Mb |      |
AZ081796 T7+,
AZ081799 SP6 | | RH53565 |
|
|
14q11.2c |
RP11-566I2 |
chr14 21Mb |      |
AZ081794 T7+,
AZ081804 SP6 | | WI-14198 |
|
|
14q11.2g~12a |
RP11-368G9 |
chr14 23Mb |      |
AZ081917 T7+,
AQ542979 T7,
AZ575482 T7,
AZ081803 SP6,
AQ542975 SP6 | | stSG41912 |
|
|
14q12a~12b |
RP11-303G24 |
chr14 25Mb |      |
AZ081787 T7+,
AZ081801 SP6 | | RH53533 |
|
|
14q12c |
RP11-529E4 |
chr14 27Mb |      |
AZ081792 T7+,
AZ081800 SP6 | | RH71202 |
|
|
14q12d |
RP11-369O9 |
chr14 29Mb |      |
AZ081947 SP6,
AZ081946 T7 | | RH47598 |
|
|
14q12e |
RP11-559L22 |
chr14 30Mb |      |
AZ081813 SP6+,
AZ081791 T7+,
AZ564736 SP6 | |
A002A15+,
A002A15 |
|
|
14q12e~13.1a |
RP11-557O15 |
chr14 32Mb |      |
AZ081978 T7,
AZ081977 SP6 | | A003N09 |
|
|
14q13.2b |
RP11-465B6 |
chr14 35Mb |      |
AZ081818 SP6+,
AZ081788 T7+,
AZ564739 SP6,
AQ581027 SP6,
AQ581028 T7 |
AL354770 | RH28319 |
|
|
14q13.2b~13.3a |
RP11-458A21 |
chr14 36Mb |      |
AZ081912 SP6+,
AZ081920 T7+,
AQ582366 T7 | | RH69670 |
|
|
14q13.3a |
RP11-26M6 |
chr14 34Mb |      |
B84609 SP6,
B84610 T7 | | SHGC-2833 |
|
|
14q21.1b |
RP11-305B23 |
chr14 38Mb |      |
AZ081823 SP6+,
AZ564740 SP6,
AZ081798 T7 | | A003B13 |
|
|
14q21.1c~21.2a |
RP11-435L2 |
chr14 42Mb |      |
AZ081817 SP6+,
AZ081810 T7,
AZ564742 SP6 | | RH26197 |
|
|
14q21.2a~21.3a |
RP11-453F20 |
chr14 44Mb |      |
AZ081812 SP6+,
AZ303305 T7,
AZ564741 SP6,
AQ587286 SP6 | | A009Y19,
RH67673 |
|
|
14q21.3c~21.3d |
RP11-315M16 |
chr14 47Mb |      |
AZ081816 SP6+,
AZ564743 SP6,
AQ504627 SP6,
AZ081811 T7,
AZ520619 T7 | | RH53701 |
|
|
14q21.3d |
RP11-321F3 |
chr14 46Mb |      |
AZ081784 SP6,
AZ081781 T7 | | RH44884 |
|
|
14q22.1b |
RP11-379K10 |
chr14 50Mb |      |
AZ081921 SP6+,
AZ081809 T7,
AQ531520 T7,
AQ531159 SP6 | | RH65693 |
|
|
14q22.1c |
RP11-368A1 |
chr14 50Mb |      |
AZ081906 T7,
AQ542780 T7,
AZ081911 SP6 | | RH26628 |
|
|
14q22.2b |
RP11-312M17 |
chr14 54Mb |      |
AZ081815 SP6+,
AZ564744 SP6,
AZ081808 T7 | | A002Q45 |
|
|
14q22.3b |
RP11-571D4 |
chr14 55Mb |      |
AZ081913 SP6+,
AZ081807 T7 | | N38942 |
|
|
14q23.1a |
RP11-571J17 |
chr14 57Mb |      |
AZ081814 SP6+,
AZ564745 SP6,
AZ081806 T7 | | A005D15 |
|
|
14q23.1b~23.1c |
RP11-471N20 |
chr14 60Mb |      |
AZ081851 T7,
AQ632875 T7,
AZ081862 SP6 | | stSG21195 |
|
|
14q23.1c |
RP11-436G5 |
chr14 61Mb |      |
AQ554747 T7+,
AZ081955 SP6,
AZ081954 T7 | | A005M45,
RH16813 |
|
|
14q23.1c~23.2a |
RP11-445J13 |
chr14 62Mb |      |
AZ081834 SP6,
AZ081833 T7 | | SGC31030 |
|
|
14q23.3a |
RP11-305I24 |
chr14 66Mb |      |
AZ081828 SP6,
AZ081827 T7 | | RH46030 |
|
|
14q24.2a |
RP11-448I10 |
chr14 69Mb |      |
AZ081930 SP6,
AZ081925 T7 | | A009C06,
RH67094 |
|
|
14q24.2a~24.2b |
RP11-325N20 |
chr14 71Mb |      |
AZ081938 SP6,
AZ081953 T7 | | RH70438 |
|
|
14q24.2b |
RP11-435K23 |
chr14 72Mb |      |
AZ081857 SP6,
AQ936166 SP6,
AQ936412 T7,
AZ081858 T7 | | RH48398 |
|
|
14q24.3c |
CTD-3211F8 |
chr14 74Mb |      |
AQ174807 T7,
AQ174955 SP6 |
AC007956 | |
|
|
14q24.3d |
RP11-501I4 |
chr14 76Mb |      |
AZ081831 T7+,
AZ081849 SP6+ |
AF111168 | RH53682 |
|
|
14q24.3d |
RP11-98L12 |
chr14 75Mb |      |
AQ320418 T7,
AQ320414 SP6 |
AC008015 | |
|
|
14q31.1b |
RP11-475O3 |
chr14 79Mb |      |
AZ081855 SP6,
AQ635862 T7,
AZ081863 T7 | | RH78382 |
|
|
14q31.1c |
RP11-477M24 |
chr14 80Mb |      |
AQ635783 T7+,
AQ635769 SP6,
AZ081845 T7,
AZ081844 SP6 | | RH44680 |
|
|
14q31.2a |
RP11-259M24 |
chr14 83Mb |      |
AQ481968 SP6,
AQ481970 T7 | | |
|
|
14q31.3c |
RP11-477I23 |
chr14 86Mb |      |
AZ081914 T7+,
AZ081918 SP6+,
AQ634805 SP6 | | RH78126 |
|
|
14q31.3d |
RP11-327L24 |
chr14 87Mb |      |
AZ081943 T7,
AZ081942 SP6 | | RH69803 |
|
|
14q32.12a~32.12b |
RP11-374H13 |
chr14 92Mb |      |
AZ081829 T7,
AZ081836 SP6 | | Cda0vd04 |
|
|
14q32.13b~32.2a |
RP11-300N18 |
chr14 93Mb |      |
AQ504718 SP6,
AZ081941 SP6,
AZ081980 T7,
AQ504720 T7 | | RH17576 |
|
|
14q32.2a |
RP11-483K13 |
chr14 95Mb |      |
AZ081904 T7,
AQ637174 T7,
AQ637172 SP6,
AZ303373 SP6 | | A001Y25,
RH10590 |
|
|
14q32.2b |
RP11-431B1 |
chr14 98Mb |      |
AZ081948 T7,
AZ081949 SP6 | | stSG35230 |
|
|
14q32.32a |
RP11-502A24 |
chr14 101Mb |      |
AZ081840 SP6+,
AZ081841 T7+ | | -,
A008V18,
RH48398+ |
|
|
14q32.32b |
RP11-454M12 |
chr14 102Mb |      |
AZ081842 SP6+,
AZ081843 T7,
AQ582808 T7 | | T86307 |
|
|
14q32.33d |
RP11-435F10 |
chr14 104Mb |      |
AZ081951 SP6,
AZ081950 T7 | | RH53818 |
1 FISH-mapped clone was placed on the sequenced
genome based on BAC-ends, insert sequences and STSs.
2 Sequences that are associated with the clone
and used for clone placement.
* indicates
sequences that disagree with data in 'Placement on genome' column.
+ indicates sequences that didn't align to the
genome or were not used for annotation.
3 Placement summary indicates sequences
used for clone placement.
 BAC-end sequence aligned in the plus orientation BAC-end sequence aligned in the plus orientation
 BAC-end sequence aligned in the minus orientation BAC-end sequence aligned in the minus orientation
 insert sequence insert sequence
 STS aligned STS aligned
 insert sequence has clone end information. insert sequence has clone end information.
|
|








