|
|
| PubMed | BLAST | OMIM | Taxonomy | Structure |
|
|
||||
|
NCBI's Structure database Short summary Direct WWW access to the MMDB server Papers about MMDB 3D-structure viewer Structure comparisons Submit structure database searches Conserved Domain Database Research topics and staff |
Index
MMDB OverviewMMDB (a Molecular Modeling Database) is Entrez's macromolecular 3D Structure database. It contains experimentally determined biopolymer structures obtained from the Protein Data Bank (PDB). Entrez provides a number of powerful search tools useful for identifying structures of interest. Starting with a particular protein sequence, for example, one can select all related sequences (as identified by BLAST) and then link to any for which 3D structure is known. This provides access to the rich information on biological function and molecular evolution that can be provided by 3D structure.MMDB's Structure Summary pages are the "home page" for each 3D structure entry. They provide a general description of the entry together with a number of links leading to further information. These include links to PubMed citations and taxonomy assignments. MMDB Structure Summary pages also provide a graphical overview of the macromolecular contents of the entry and the biological annotation available for each. Lastly, Structure Summaries provide links to 3D visualization.
Structure DescriptionsDescription information in MMDB comes largely from PDB. It includes:
Graphical SummariesThe graphical display in a MMDB summary page lists macromolecular contents of the structure entry and the biological annotation available for each chain.The image below illustrates 4 compact 3D domains identified in chain A and B, respectively, and 4 nucleotide chains (C, D, E and F) in the MMDB entry 1RUO (left) and graphics in MMDB structure summary page (right). Links to the Conserved Domain Database are also shown. 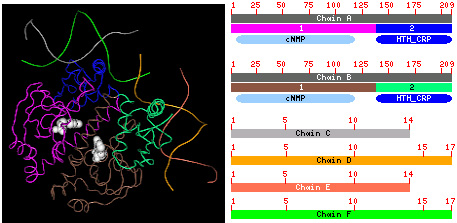
The 3D structure is viewed by Cn3D. The protein chains (A and B) bind to nucleotide chains (C, D, E and F) and a small substrate cNMP (white filling space ball). In the graphical display, each chain is shown as a sequence-ruler with label. The colored boxes with number below the ruler indicate locations of 3D domains in each protein sequence. If one domain is split into several pieces by geometric criteria, these pieces will have the same color. The color is cycled in the same order employed in Cn3D with coloring option "Domain" against both protein and nucleotide chains, according to the domain numbers. Clicking these boxes will display the structural neighbors of each compact domain in a VAST page. At the left of the sequence ruler, a hotlink named "protein" or "nucleotide" (not shown here) goes to Entrez sequence browser, where you can get information about the sequence itself. And clicking the link "3D Domain" (not shown here), which locates below the "protein" link, will go to Entrez domain browser to display the information related to each domain in the structure. Links to the Conserved Domain Database (CDD) name and describe recurrent evolution modules. They are computed using RPS-Blast, which can be explored in CD-search, and are displayed as ovals of various colors. Each oval represents a cluster of related domain models where at least one scored above the RPS-Blast significance threshold of e <0.1. Overlaping hits are clustered if the mutual overlap between them are greater than 50%. In a one-against-one comparison, if the average overlap of a pair of overlapping representatives is greater than 50%, the less similar one to the query will not be displayed. The color is cycled in a similar order as in CDART. Each oval has a brief CD name on it, if the space is big enough, and clicking it will go to the corresponding CD multiple alignment browser to display how the chain is aligned with the sequences included in the CD. The link named "CDs" at the left of CD rectangles goes to Entrez CD browser.
Viewing 3D StructureMMDB Structure Summary pages support three different structure viewers: Cn3D, an Entrez helper application developed by NCBI, RasMol and MAGE. All three are available for a variety of platforms (Windows, MacOS and Unix). To be able to visualize 3D structures, you must download one of these viewers and install it on your local machine. Usually the web-browser needs to be configured too. Click here for further instructions.By selecting from three pop-up menus and clicking the "View / Save" button, you may launch a structure viewer.
The first menu box should be set up to "Structure" in order to display a 3D structure. From the second menu, you may select one of the three viewers for structural visualization. NCBI's Cn3D viewer is the default. MMDB structure data contains different "models". A "model" is the 3D representation of a structure, containing explicit 3D coordinates. There are different levels of "structure complexity". You may choose from:
In Cn3D, you will be able to toggle off and on backbone and sidechain representations for the individual chains in the structure, as well as choose between different levels of complexity in the backbone representation. Cn3D displays information on metals and ionic elements, such as those found in metalloproteins, and highlights them accordingly.
Saving Structure DataYou may look at a structure file in your web-browser, instead of launching a viewer for the respective format, or save it as a disk file. This permits you to load the structure into a viewer later. To save a structure, select "Save File" from the menu and "Cn3D" on other option and then press "View / Save". A request for a file name will be prompted.
The option of "See File" in the first menu box will display the structure data on the browser, should one want to examine the numeral data.
What are "3D Domains" and "Conserved Domains"?3D domains within individual polypeptide chains in MMDB are identified automatically, using an algorithm that searches for one or more breakpoints, falling between major secondary structure elements, such that the ratio of intra- to inter-domain contacts falls above a set threshold. The 3D domains identified in this way provide means to increase the sensitivity of structure neighbor calculations, and to present 3D superpositions based on compact domains as well as on complete polypeptide chains. They are not intended to represent domains identified by comparative sequence and structure analysis, as modules that recur in related proteins, though there is often good agreement between domain boundaries identified by these methods. The structure similarities among individual chains and their compact 3D domains in MMDB are calculated by VAST algorithm, which superposes structures based on the structure alignments of their secondary structure elements. Conserved domains are recurring units (sequence and structure motifs) in molecular evolution whose extents have been determined by comparative analysis. Molecular evolution readily uses such domains as building blocks which may be recombined in different arrangements. NCBI's Conserved Domain Database (CDD) provides detailed information for such domains, including descriptions, citations and multiple alignments.
"Obsolete" StructuresSuch a entry is available in MMDB but not indexed in entrez. The reason is that either it was replaced by a new entry or withdrawn from PDB. If the entry is indicated by "replaced by", then clicking the number following the words will launch a MMDB summary page for the new entry. |
||
|
Updated 11/28/02 |
|
| Help Desk | NCBI | NLM | NIH | Credits |