|
January 2003
![]()
The genus Listeria contains 6 species: L. monocytogenes, L. innocua, L. seeligeri, L. welshimeri, L. ivanovii, and L. grayi (Table 1). L. grayi (29, 34) and L. ivanovii (12, 28) each contain two subspecies, which do not need to be specified in this analysis. A recent taxonomic review of the genus by Rocourt (35) updates the previous reviews (11, 37). L. ivanovii and L. monocytogenesare pathogenic for mice and other animals. However, only L. monocytogenes is commonly associated with human listeriosis. Listeriosis associated infection by L. ivanovii, and even L. seeligeri, is extremely rare in humans. The universal occurrence of L. monocytogenes in food (36) and the risk of contracting food-borne L. monocytogenes listeriosis (41b, http://www.foodsafety.gov/~dms/lmrisk.html) have been thoroughly reviewed recently. This chapter describes the detection and enumeration of L. monocytogenes in foods. Detection of this pathogen in the food processing environment, such as on food contact surfaces and equipment, is described elsewhere (41a).
The preferred standard methodology, and permitted alternative rapid methodologies, to be used for detection and isolation of Listeria monocytogenes are as follows. Presumptive contaminated food lots are sampled. Generally, sub-samples are composited if required by FDA field laboratory instructions. Analytical portions (25 g) are pre-enriched for Listeria species at 30° C for 4 h in buffered Listeria enrichment broth (BLEB), equivalent (26) to AOAC/IDF dairy products enrichment broth (4, 40) base containing sodium pyruvate (43). At the fourth hour of the incubation, the selective agents (acriflavin, 10 mg/L (4, 40); sodium nalidixate, 40 mg/L; optional antifungal, e.g. cycloheximide 50 mg/L) are added. Incubation for selective enrichment is continued at 30° C for a total of 48 h. The enrichment culture is streaked at 24 and 48 h on one of the prescribed differential selective-agars in order to isolate Listeria species.
Alternatively, prescribed rapid detection kits with their respective enrichment media may be conditionally used to screen for presence of Listeria contaminants. Putative listeria isolates on selective agars from standard or screen positive enrichments are purified on non-selective agar and confirmed by conventional identification tests or by a battery of such tests in kit form. Isolates may be rapidly confirmed as L. monocytogenes (or not) by using specific test kits. Subtyping of L. monocytogenes isolates is optional except for FDA isolates, which have to be typed serologically, and by pulsed-field gel electrophoresis (PFGE) and by ribotyping. Non-obligatory pathogenicity testing of L. monocytogenes isolates is described.
Enumeration of L. monocytogenes in positive samples is performed on reserve sample by colony count on L. monocytogenes differential selective agar in conjunction with MPN enumeration using selective enrichment in BLEB with subsequent plating on ALOA or BCM differential selective agar.
The major revisions to the Listeria methodology follow.
Certain prescribed rapid detection kits and their enrichments are now authorized screening alternatives to the standard selective enrichment.
It is now necessary to use only one instead of two of the several prescribed selective isolation agars (Oxford agar, PALCAM, LPM plus esculin and ferric iron, MOX). Oxford agar is still the preferred standard selective isolation medium. MOX has been added to the list of prescribed selective agars and LPM without added esculin and ferric iron has been removed. Trial use of the new chromogenic differential selective agars, like BCM, ALOA, CHROMagar Listeria and Rapid' L. mono, is encouraged as long as it is in parallel with one of the prescribed selective agars. The new agar media differentiate L. monocytogenes/L. ivanovii colonies from those of other Listeria spp. and will greatly facilitating picking of L. monocytogenes colonies when colonies of more than one species are present on a plate.
The Henry illumination technique is de-emphasized because only differential selective isolation agars are prescribed.
The current enrichment medium, which resembles the step-1 enrichment of the internationally harmonized method proposed by Asperger et al. (10a), is basically unchanged. However, pimaricin (natamycin), a much less toxic compound than cycloheximide, is introduced as the alternative antifungal compound in the Listeria enrichment medium,
If L. monocytogenes is detected in a food sample, enumeration of the level of contamination in the food is required.
| Acid produced from | |||||
|---|---|---|---|---|---|
| Species | Mannitol | Rhamnose | Xylose | Virulenceb | |
| L. monocytogenes | + | - | + | - | + |
| L. ivanoviic | + | - | - | + | + |
| L. innocua | - | - | Vd | - | - |
| L. welshimeri | - | - | Vd | + | - |
| L. seeligeri | + | - | - | + | - |
| L. grayie | - | + | Vd | - | - |
| a Sheep blood agar stab. | |||||
| b Mouse test. | |||||
| c Ribose fermenting strains are classified as L. ivanovii subsp. ivanovii and ribose non-fermenters as L. ivanovii subsp. londiniensis. | |||||
| d V, variable biotypes | |||||
| e Includes two subspecies - L. grayi subsp. murrayi reduces nitrate L. grayi subsp. grayi does not reduce nitrate. | |||||
Equipment and materials
Note: Alternative companies may be used when the products are equivalent.
Sampling and Enrichment Procedures
Sample treatment. Sample refrigeration at 4°C is recommended for handling, storing, and shipping materials to be analyzed for L. monocytogenes, which will grow, although slowly, at this temperature if other conditions permit. However, if the sample is already frozen, it should not be thawed until analysis.
Composited samples. Generally, composited samples are prepared as follows. A food lot sample is collected consisting of 10 sub-samples (liquid, cream or solid food) and 50-g or ml portions of each sub-sample are used to make two composite samples (250 g each). Take care to make sub-samples representative of a food's outer surface as well as its interior. For the first composite 5 x 50-g portions from 5 sub-samples are pooled and blended or stomached in 250 ml buffered Listeria enrichment broth base containing sodium pyruvate without selective agents (BLEB, M52). The second composite is made from the remaining 5 sub-samples in the same way. Both blended composites contain 250-g food portions and 250 ml basal BLEB.
Normally 50 g of each composite blend (equivalent to 25g food plus 25 ml basal BLEB) is mixed with a 200 ml amount of basal BLEB. Since there are two composites, the end result is two 25-g analytical portions each contained in 225 ml amounts of basal BLEB. Thus, for each sub-sample a total of 50 g of composited food is analyzed. An aliquot (50 ml) of the composite blend should be retained, preferably at 5° C and not below 0° C, for possible pathogen enumeration.
Non-composited samples. If composite samples are not required, single 25-g analytical portions of food are simply blended or stomached in 225 ml of basal BLEB and pre-enriched/enriched as described later. A 25-g sample should be retained for possible pathogen enumeration. Store it at 5° C if it is not frozen or, if frozen, in a non-defrosting freezer.
Pre-enrichment and enrichment. Incubate for 4 h at 30° C, add the selective agents and continue incubating for a total time of 48 h at 30° C. If cycloheximide is unavailable, the preferred substitute is pimaricin (natamycin) at 25 mg/L (27). Natamycin is much safer to use than cycloheximide. Another possibility, if the matrices of interest (e.g. pasteurized milk and cream, yogurt, and precooked frozen seafood) are low in yeast and mold, is to do without an anti-fungal agent. This is not advisable for mold-ripened cheeses, smoked or dried seafood or fresh produce.
Enrichment with enumeration. Surveillance enumeration of Listeria monocytogenes levels in contaminated food is now required. Detection may be done first and if contamination is detected, a reserve sample portion can be enumerated. This is probably the preferable method as, generally, only a few percent of samples can be expected to be positive and then most often at a level of only about 1 cfu/25g. However, the option of combining regulatory detection and enumeration is provided in Enumeration.
Prescribed Alternative Methods for Screening Enriched Samples. For the prescribed methods kits listed in Table 2, follow the manufacturers' package insert instructions making certain they have not deviated from the approved versions of the AOAC INTERNATIONAL Official Methods Manual protocols (Table 2). The kits are only approved for the specified food matrices, listed in Chapter 10, Supplement 1A, which vary from kit to kit. For other food matrices listed in Chapter 10, Supplement 1B, in-house validation is necessary. The easiest way to validate is to streak all kit-associated 48-h enrichments, which give kit-negative results, on a plate of one of the prescribed standard method esculin agars. Even with the validated matrices the 48-h streaking of kit-negative enrichments to esculin selective agars is recommended for five or so different lots of a given food matrix until the analyst is confident of the kit's performance with that particular matrix. This is because conventional validation of matrices does not take into account qualitative and quantitative competitive microflora variation among different lots of a given food matrix. Strong competition will impact rapid kit methods more than cultural methods. Kit thresholds of detection (>104 cfu/ml of enrichment culture) are higher than that of the culture plate method (about 102 cfu/ml of enrichment culture) so that, with very competitive microflora, kits could give false negative results. Alternatively, the limit of detection (50% endpoint) by quantitative spiking with a single strain of L. monocytogenes should be determined. It should be not more than 3 cfu per 25-g analytical portion. A value greater than this would only be acceptable if the conventional culture method's performance level is comparable. For all food matrices, a kit positive result must be supported by a culture of the L. monocytogenes isolate.
Table 2. Listeria Genus Detection Test Kits Prescribed for Regulatory Screening
AOAC Official Method 993.09. 2000. Listeria in dairy products, seafoods, and meats. Colorimetric deoxyribonucleic acid hybridization method (GENE-TRAK Listeria Assay). (3, 14)
AOAC Official Method 994.03. 2000. Listeria monocytogenes in dairy products, seafoods, and meats. Colorimetric monoclonal enzyme-linked immunosorbent assay method (Listeria Tek). (5, 15, 32)
AOAC Official Method 995.22. 2000. Listeria in foods. Colorimetric polyclonal enzyme immunoassay screening method (TECRA Listeria Visual Immunoassay [TLVIA]). (6, 30)
AOAC Official Method 996.14. 2000. Assurance (Polyclonal Enzyme Immunoassay Method). (7, 21)
AOAC Official Method 997.03. 2000. Visual Immunoprecipitate Assay (VIP). (8, 22)
AOAC Official Method 999.06. 2000. Enzyme Linked Immunofluorescent Assay (ELFA) VIDAS LIS Assay Screening Method. (9, 23)
Isolation procedure
At 24 and 48 h, streak BLEB culture onto one of the following esculin-containing selective isolation agars: either OXA (16, M118) or PALCAM (42, M118a) or MOX (41, M103a) or LPM (31, M81) fortified with esculin and Fe3+ (M82). These esculin-containing media are listed in order of preferred use, subject to their availability. Incubate OXA, PALCAM or MOX plates at 35° C for 24-48 h and fortified LPM plates at 30° C for 24-48 h. It is strongly recommended that one of the L. monocytogenes-L. ivanovii differential selective agars, such as BCM (33, M117a), ALOA (M10a), RapidL'mono (M131a), or CHROMagar Listeria (M40a) be streaked at 48 h (optionally at 24 h, too) in addition to the chosen esculin-containing selective agar. This will reduce the problem of masking of L. monocytogenes by L. innocua. [Note: BCM has been collaboratively validated by FDA (26a). An ISO TC34 SC9 comparative validation showed that all the media (and a selective blood agar - LMBA, Sifin, Germany) inhibited Listeria competitors more or less equally well. ALOA was preferred only because its formulation is public. Another differential selective medium, Chromogenic Listeria Agar (M40b) is due to be marketed in the future.]
Listeria colonies are black with a black halo on esculin-containing media. Certain other bacteria can form weakly brownish black colonies, but color development takes longer than 2 days. Transfer 5 or more typical colonies from OXA and PALCAM or modified LPM or MOX to Trypticase soy agar with yeast extract (TSAye), streaking for purity and typical isolated colonies. If BCM plates are streaked as recommended above and blue colonies are observed, they are presumptive L. monocytogenes colonies since L. ivanovii is not often reported in foods. L. monocytogenes and L. ivanovii colonies on ALOA are blue and have a zone of lipolysis around them. Purification on TSAye is a mandatory step in the conventional analysis because isolated colonies on selective agar media may still be in contact with an invisible weak background of partially inhibited competitors. At least 5 isolates are necessary because more than one species of Listeria may be isolated from the same sample. Use of BCM and ALOA plates will help to reduce the number of colonies that need to be picked. L. monocytogenes and L. ivanovii can be distinguished using a commercial Confirmatory Medium (Biosynth International, Inc.) or by conventional rhamnose/xylose fermentation broths or agars. Incubate TSAye plates at 30° C for 24-48 h. The plates may be incubated at 35° C if colonies will not be used for wet-mount motility observations (see E-2, below). For the approved rapid methods (Table 2), use the selective isolation agar recommended by the manufacturer but, as noted above, auxiliary use of the new L. monocytogenes-L.ivanovii differential agars is also recommended.
Identification procedure
Identify purified isolates by the following classical tests (E, 1-11). Rapid kits are available to facilitate biochemical testing to genus or species level (see E-11 and E-12).
Examine TSAye plates for typical colonies. Observation with Henry oblique transmitted illumination can be helpful at this stage but is not mandatory (See ref. 25 for details).
Pick typical colony from culture plate incubated at 30°C or less and examine by wet mount, using 0.85% saline for suspending medium and oil immersion objective of phase-contrast microscope. Choose a colony with enough growth to make a fairly heavy suspension; emulsify thoroughly. If too little growth is used, the few cells present will stick to the glass slide and appear non-motile. Listeria spp. are slim, short rods with slight rotating or tumbling motility. Always compare with known culture. Cocci, large rods, or rods with rapid, swimming motility are not Listeria spp. Alternatively, use the 7-day motility test medium (see E-9).
Test typical colony for catalase. Listeria species are catalase-positive.
Gram stain 16- to 24-h cultures. All Listeria spp. are short, Gram-positive rods; however, with older cultures the Gram stain reaction can be variable and also cells may appear coccoidal. The cells have a tendency to palisade in thick-stained smears. This can lead to false rejection as a diphtheroid.
Pick typical colony to a tube of TSBye for inoculating carbohydrate fermentation and other test media. Incubate at 35° C for 24 h. This culture may be kept at 4°C several days and used repeatedly as inoculum. Commercial kits are available for isolate identification (see E-11)
Inoculate heavily (from TSAye colony) 5% sheep blood agar by stabbing plates that have been poured thick and dried well (check for moisture before using). Draw grid of 20-25 spaces on plate bottom. Stab one culture per grid space. Always stab positive controls (L. ivanovii and L. monocytogenes) and negative control (L. innocua). Incubate for 24-48 h at 35° C. Attempt to stab as near to bottom of agar layer as possible, without actually touching bottom of agar layer and possibly fracturing the agar.
Examine blood agar plates containing culture stabs with bright light. L.monocytogenes and L. seeligeri produce a slightly cleared zone around the stab. L. innocua shows no zone of hemolysis, whereas L. ivanovii produces a well-defined clear zone around the stab. Do not try to differentiate species at this point, but note nature of hemolytic reaction. Resolve questionable reactions by the CAMP test. (Note: Hemolysis is more easily determined when the depth of the blood agar is thinner than the usual 5mm. Optionally, this may be achieved by use of a blood agar overlay (1-2 mm) technique).
Nitrate reduction test. This test is optional. Only L. grayi ssp. murrayi reduces nitrates. The test distinguishes L. grayi ssp. murrayi from L. grayi ssp. grayi. Use a TSBye culture to inoculate nitrate broth (M108). Incubate at 35° C for 5 days. Add 0.2 ml reagent A, followed by 0.2 ml reagent B (R48). A red-violet color indicates presence of nitrite, i.e. nitrate has been reduced. If no color develops, add powdered zinc and hold for 1 h. A developing red-violet color indicates that nitrate is still present and has not been reduced.
As an alternative procedure (R48), add 0.2 ml reagent A followed by 0.2 ml reagent C. An orange color indicates reduction of nitrate. If no color develops, add powdered zinc as above. Development of an orange color indicates unreduced nitrate.
Inoculate SIM or MTM from TSBye. Incubate for 7 days at room temperature. Observe daily. Listeria spp. are motile, giving a typical umbrella-like growth pattern. MTM provides the best defined umbrellas. Alternatively, observe the 30° C TSBye cultures, by phase contrast microscopy (x1000) for tumbling motility.
From TSBye culture, inoculate the following carbohydrates as 0.5% solutions in purple carbohydrate broth (the use of Durham tubes is optional): dextrose, esculin, maltose, rhamnose, mannitol, and xylose. Incubate 7 days at 35° C. Positively reacting Listeria spp. produce acid with no gas. Consult Table 1 for xylose-rhamnose reactions of Listeria spp. All species should be positive for dextrose, esculin, and maltose. All Listeria spp. except L. grayi should be mannitol-negative. If pigmentation of the isolate on OXA, PALCAM, MOX or LPM plus esculin/Fe3+ is unequivocal, the esculin test may be omitted.
Purified isolates may be rapidly identified by using commercial kits (additional tests may be needed to speciate completely): Vitek Automicrobic Gram Positive and Gram Negative Identification cards (bioMerieux, Hazelwood, MO) or API Listeria (bioMerieux, Marcy-l'Etoile, France) which does not require an additional CAMP test. The MICRO-IDTM kit (bioMerieux, Hazelwood, MO; 1, 24) permits speciation of Listeria isolates if their CAMP reactions are known. The Phenotype MicroArray for Listeria (BiOLOG, Hayward, CA) is another recently introduced kit.
Alternative rapid methods that are prescribed for identifying Listeria isolates as L. monocytogenes are listed in Table 3. Depending on the kit, isolates may be identified in pure culture or from OXA or the other selective isolation agars. Purified isolates identified as Listeria monocytogenes by these tests should be retained for regulatory reference.
Table 3. Test Kits Useful in Confirming Listeria Isolates as Listeria monocytogenes or not*
AccuProbeTM Listeria monocytogenes culture confirmation test (Gen-Probe, Inc, San Diego, CA; 10, 19).
GeneTrak Listeria monocytogenes test kit (Neogen, Lansing, MI; 19).
Probelia Listeria monocytogenes test kit (BioControl, Seattle, WA).
VIDAS Listeria monocytogenes test kit (bioMerieux).
Transia Plate Listeria monocytogenes (Diffchamb SA, Lyon, France)(38)
FDA, SRL application of Niederhauser et al. method for PCR detection and identification of L. monocytogenes (32a, 32b)
BAX Listeria monocytogenes test. (Qualicon, Inc., Wilmington, DE) (9a).
* These kits are in various stages of validation and when suitably validated can also be used to screen enrichments for L. monocytogenes. Presently, FDA only prescribes validated kits that screen for all Listeria species.
The CAMP Test.
The Christie-Atkins-Munch-Peterson (CAMP) test (Table 4 and Fig. 1) is useful in confirming species
particularly when blood agar stab test results are equivocal. To perform the test, streak a
![]() -hemolytic
Staphylococcus aureus and a Rhodococcus equi culture in parallel and diametrically opposite
each other on a sheep blood agar plate. Streak several test cultures parallel to one another, but
at right angles to and between the S. aureus and R. equi streaks. After incubation
at 35° C for 24-48 h, examine the plates for hemolysis. L. monocytogenes and L. seeligeri
hemolytic reactions are enhanced in the zone influenced by the S. aureus streak. The other
species remain non-hemolytic. The L. monocytogenes reaction is often optimal at 24 h rather
than 48 h. To obtain enough R. equi to provide a good streak of growth, incubate the slant
culture 48 h rather than 24 h. Use of known control isolates of Listeria spp. on a separate
sheep blood agar plate is recommended. Sheep blood agar plates should be as fresh as possible.
-hemolytic
Staphylococcus aureus and a Rhodococcus equi culture in parallel and diametrically opposite
each other on a sheep blood agar plate. Streak several test cultures parallel to one another, but
at right angles to and between the S. aureus and R. equi streaks. After incubation
at 35° C for 24-48 h, examine the plates for hemolysis. L. monocytogenes and L. seeligeri
hemolytic reactions are enhanced in the zone influenced by the S. aureus streak. The other
species remain non-hemolytic. The L. monocytogenes reaction is often optimal at 24 h rather
than 48 h. To obtain enough R. equi to provide a good streak of growth, incubate the slant
culture 48 h rather than 24 h. Use of known control isolates of Listeria spp. on a separate
sheep blood agar plate is recommended. Sheep blood agar plates should be as fresh as possible.
Hemolysis enhancement with |
||
|---|---|---|
| Staphylococcus aureus (S) | Rhodococcus equi (R) | |
| L. monocytogenes | + | -* |
| L. ivanovii | - | + |
| L. innocua | - | - |
| L. welshimeri | - | - |
| L. seeligeri | + | - |
| * Rare strains are S+ and R+. The R+ reaction is less pronounced than that of L. ivanovii. | ||
| CAMP test strains are available from culture collections, including the American Type Culture Collection (ATCC), Manassas, VA http://www.atcc.org | ||
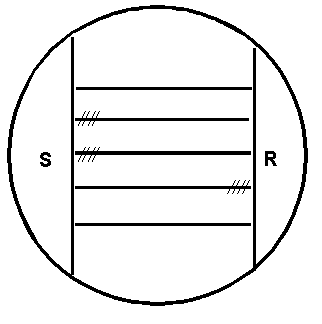
Figure 1. CAMP test for Listeria monocytogenes: Inoculation pattern of the sheep blood agar plate. Horizontal lines represent streak inoculations of 5 test strains. Vertical lines represent streak inoculations of Staphylococcus aureus (S) and Rhodococcus equi (R). Hatched lines indicate (diagrammatically only) the locations of hemolysis enhancement regions.
Streak weakly ![]() -hemolytic S. aureus
FDA strain ATCC 49444 (CIP 5710; NCTC 7428) or strains ATCC 25923 and R. equi (ATCC 6939; NCTC
1621) vertically on sheep blood agar. Separate vertical streaks so that test strains may be streaked
horizontally between them without quite touching them. After 24- and 48-h incubation at 35° C,
examine plates for hemolysis in the zone of influence of the vertical streaks. Figure 1 shows the
arrangement of the culture streaks on a CAMP plate. Hemolysis of L. monocytogenes and L.
seeligeri is enhanced near the S. aureus streak; L. ivanovii hemolysis is enhanced
near the R. equi streak. Other species are non-hemolytic and do not react in this test. The
CAMP test differentiates L. ivanovii from L. seeligeri and can differentiate a weakly
hemolytic L. seeligeri (that may have been read as non-hemolytic) from L. welshimeri.
Isolates giving reactions typical for L. monocytogenes except for hemolysis should be CAMP-tested
before they are characterizedas L. innocua. A factor easily prepared from S. aureus
cultures can be used to enhance hemolysis by L. monocytogenes and L. seeligeri in sheep
blood agar plates. Disks impregnated with the
-hemolytic S. aureus
FDA strain ATCC 49444 (CIP 5710; NCTC 7428) or strains ATCC 25923 and R. equi (ATCC 6939; NCTC
1621) vertically on sheep blood agar. Separate vertical streaks so that test strains may be streaked
horizontally between them without quite touching them. After 24- and 48-h incubation at 35° C,
examine plates for hemolysis in the zone of influence of the vertical streaks. Figure 1 shows the
arrangement of the culture streaks on a CAMP plate. Hemolysis of L. monocytogenes and L.
seeligeri is enhanced near the S. aureus streak; L. ivanovii hemolysis is enhanced
near the R. equi streak. Other species are non-hemolytic and do not react in this test. The
CAMP test differentiates L. ivanovii from L. seeligeri and can differentiate a weakly
hemolytic L. seeligeri (that may have been read as non-hemolytic) from L. welshimeri.
Isolates giving reactions typical for L. monocytogenes except for hemolysis should be CAMP-tested
before they are characterizedas L. innocua. A factor easily prepared from S. aureus
cultures can be used to enhance hemolysis by L. monocytogenes and L. seeligeri in sheep
blood agar plates. Disks impregnated with the ![]() -lysin of S. aureus (REMEL, Lenexa, KS) can be used for the same purpose.
-lysin of S. aureus (REMEL, Lenexa, KS) can be used for the same purpose.
Subtyping of L. monocytogenes isolates (Required)
FDA isolates should be typed serologically and genetically.
Serological typing. Use commercial sera to characterize isolates as type 1, type 4 or not type 1 or 4 (types 3, 5, 6 etc.).
| Listeria species | Serotypes |
|---|---|
| L. monocytogenes | 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4ab, 4b 4c, 4d, 4e, "7" |
| L. ivanovii | 5 |
| L. innocua | 4ab, 6a, 6b, Una |
| L. welshimeri | 6a, 6b |
| L. seeligeri | 1/2b, 4c, 4d, 6b, Un |
| a Un, undefined | |
Table 5 exhibits the serological relationships of Listeria spp. Most L. monocytogenes isolates obtained from patients and the environment are type 1 or 4. More than 90% of L. monocytogenes isolates can be serotyped with commercially available sera. All nonpathogenic species, except L. welshimeri, share one or more somatic antigens with L. monocytogenes. Serotyping alone without thorough characterization, therefore, is not adequate for identification of L. monocytogenes.
Serology is useful when epidemiological considerations are crucial. Use a TSBye culture to inoculate Tryptose broth. Incubate for 24 h at 35° C, at which temperature flagella (H) antigen expression is reduced. Transfer to Tryptose agar slants and incubate for 24 h at 35° C. Wash both slants in a total of 3 ml Difco fluorescent antibody (FA) buffer and transfer to a sterile 16 x 125-mm screw-cap tube. Heat in a water bath at 80°C for 1 h. Sediment cells by centrifugation at 1600 g for 30 min. Remove 2.2-2.3 ml of supernatant fluid and resuspend the pellet in the remainder of buffer. Follow manufacturer's recommendations for sera dilution and agglutination procedure. If flagella (H) or somatic (O) sub-factor serotyping is desired, see Chapter 11.
Genetic subtyping. Data from pulsed-field gel electrophoresis (PFGE) of DNA restriction fragments of FDA isolates should be submitted to PulseNet (CDC, Atlanta, GA). Isolates should also be ribotyped or sent to a ribotyping laboratory.
Immunocompromised Mouse Pathogenicity Test (optional).
The classical tests for Listeria pathogenicity are the Anton conjunctivitis test (rabbits), inoculation of mice, and inoculation of embryonated eggs. The immunocompromised mouse test, using intra-peritoneal (i.p.) injection, used here, is recommended because of its greatly improved sensitivity (39). Confirmation of L. monocytogenes animal pathogenicity is not needed for clinical isolates and is optional for food isolates. An isolate should be identified as L. monocytogenes if it meets all the other criteria outlined in this chapter.
About 4 mg of carrageenan (Sigma type II) dissolved in distilled water (40 mg/ml) is injected, i.p., into 18-20 g mice 24 h before the Listeria challenge. The exact volume of carrageenan solution injected, about 0.1 ml, depends on the mouse body weight. The target dose is 200 mg carrageenan per kg bodyweight. Grow isolate for 24 h at 35° C in TSBye. Transfer to 2 tubes of TSBye for another 24 h at 35° C. Place a total of 10 ml culture broth from both tubes into a 16 x 125 mm tube and centrifuge at 1600 g for 30 min. Discard supernatant and resuspend pellet in 1 ml of phosphate buffered saline. This suspension will contain approximately 1010 bacteria/ml; dilute to 105 bacteria per ml and determine actual concentration by a pour or spread-plate count. Inject (i.p.) 16 to 18 g immunocompromised Swiss white mice (5 mice/culture) with 0.1 ml of the concentrated suspension, i.e. approximately 104 bacteria per mouse. Observe for death over 5-day period. Nonpathogenic strains will not kill, but 104 pathogenic cells will kill, usually within 3 days. Use known pathogenic and nonpathogenic strains and carrageenan-treated, uninoculated mice as controls. Use 5 mice per control group. Carrageenan controls should be challenged with 0.1 ml PBS.
Interpretation of test data
The importance of completely characterizing each isolate cannot be overemphasized. Partial characterization, even if accurate, may be misleading. Since all Listeria species test negative for indole, oxidase, urease, and H2S production from organic sulfur compounds (H2S is produced from thiosulfate in the MICRO-ID test kit) and test positive for methyl red and Voges-Proskauer, these tests are discretionary. Brochothrix, which is closely related phylogenetically to Listeria, is distinguishable from Listeria by its inability to grow at 35° C and by its lack of motility. Distinguishing features of the Gram-positive non-sporeforming rods, Erysipelothrix and Kurthia, which occur rarely in Listeria analysis, can be found elsewhere (11, 37).
All Listeria spp. are small, catalase-positive, Gram-positive rods that are motile in wet mounts and in SIM. They utilize dextrose, esculin, and maltose, and some species utilize mannitol, rhamnose, and xylose with production of acid. An isolate utilizing mannitol with acid production is L. grayi. L. monocytogenes, L. ivanovii, and L. seeligeri produce hemolysis in sheep blood stabs and consequently are CAMP test-positive. Of the three, only L. monocytogenes fails to utilize xylose and is positive for rhamnose utilization. The difficulty in differentiating L. ivanovii from L. seeligeri can be resolved by the CAMP test. L. seeligeri shows enhanced hemolysis at the S. aureus streak. L. ivanovii shows enhanced hemolysis at the R. equi streak. Of the non-hemolytic species, L. innocua may provide the same rhamnose-xylose reactions as L. monocytogenes but it is negative in the CAMP test. L. innocua sometimes gives negative results for utilization of rhamnose . The significance of the undocumented reference (41) to hemolytic L. innocua isolates is unclear since it is commonly accepted that L. innocua is non-hemolytic and L. monocytogenes is hemolytic. A L. welshimeri isolate that is rhamnose-negative may be confused with a weakly hemolytic L. seeligeri isolate unless resolved by the CAMP test. Sometimes aberrant listeria strains are isolated which are extremely difficult to speciate (27a). (See Guideline for BAM Users on Identification of Atypical Hemolytic Listeria Isolates.) If such an aberrant Listeria isolate is obtained, contact [Note: The clinical significance of a strain of L. monocytogenes that is phenotypically hemolytic-negative is debatable. If it is due to a defect of the hemolysin gene, especially a deletion rather than a point mutation, it is likely clinically less significant than a normal strain would be, judging from laboratory studies of constructed hemolysin mutants in mice. However, if it is due to a regulatory defect that affects the expression of the hemolysin gene in vitro, the possibility of conditional expression in vivo is raised. Until convenient methods are devised to distinguish these structural and regulatory alternatives, the isolate need only be carefully confirmed as being a strain of L. monocytogenes phenotypically hemolysin-negative in vitro so that a soundly based regulatory decision can then be made, based upon all the relevant circumstances.]
Only after all other results are available does serotyping and other kinds of typing of Listeria isolates become meaningful. Biochemical, serological and pathogenicity data are summarized in Tables 1, 4 and 5. All data collection must be completed before species identities are determined. FDA no longer conducts routine bacteriophage-susceptibility typing of L. monocytogenes isolates.
Enumeration (required)
If a sample tests positive for L. monocytogenes, use a reserve portion of sample for enumeration. Current methods of enumeration are only presumptive for Listeria monocytogenes and some degree of further testing of isolated Listeria colonies is necessary. Conventional enumeration is described and alternative rapid methods are indicated. The proportion of presumptive isolates that are actually L. monocytogenes may be determined by conventional or rapid tests. Flexibility in choice of methods and adaptations of them is permitted but the observed count must be reported with 95% confidence limits, the method used named and any modifications indicated. The correction factor for converting the observed count to L. monocytogenes numbers must be reported as the whole number ratio of number of isolates identified as L. monocytogenes to the total number of Listeria isolates tested.
All enumeration methods, including microscopic, colony and Most Probable Number (MPN) counts are fundamentally governed by the Poisson distribution law of infrequent events. This describes the distribution of Listeria among the arrays of compartments (tubes, wells, counting chamber squares, filter grid squares, and virtual squares on culture agar surfaces). Compartmentalization separates or delineates colony-forming units in the various methods. In general, the confidence limits (CLs) of these estimates are considered proportional to the square root of the observed count. [The tabulated CLs for MPN results are asymmetric about the mean because they are usually obtained with low numbers of tubes (3 or 5) near the dilution endpoint.] As the count increases its confidence limits, expressed as a percentage of the count, decrease. Thus, choosing among methods largely reduces to a consideration of material and labor expenses and to how inoculation manipulations for an optimal number of compartments can be reduced by techniques such as filtration, semi-automation and robotics.
Surveillance Enumeration. This is required for accumulating data on cell numbers of L. monocytogenes in regulatory samples that test positive for the pathogen. Contact if you are unable to enumerate the reserve portion of a positive sample in order to try to arrange for its enumeration. To estimate the degree of sample contamination by presumptive L. monocytogenes, quantify the initial enrichment broth, before starting incubation, by direct spread plate count on ALOA, BCM or equivalent differential agar. Also, use a 3 or more-tube/well MPN culture procedure on 1, 0.1, 0.01and 0.001-g samples in BLEB (30° C, 48 h, with or without pyruvate and without delayed addition of selective agents) followed by streaking on the chosen selective agar. If all the MPN tubes are Listeria positive, use reserve sample to repeat the MPN determination using an appropriate range of more dilute analytical portions, e.g. 10-4, 10-5, 10-6, 10-7, and 10-8 g.
If selective agar plates are in short supply, an economic alternative to spreading dilution aliquots on individual selective agar plates is the drop plating method. Using a multi-channel pipette is well suited to this method. Decimally dilute 10 µl amounts of the contents of the enrichment containers in 90 µl amounts of TSBye in micro-titer plates with round-bottomed wells. Mix with a gentle circular motion of the micropipette tip before changing the tip for the next dilution. Carefully plate 10 µl of the dilutions as drops on plates of ALOA, BCM or equivalent agar. Let the droplets be absorbed before inverting the plates for incubation. Square plates are most convenient and efficient for this technique.
| Method | Reference | Validation | Specificity Matrix |
|---|---|---|---|
| MPN filter | Entis & Lerner (20) | AOAC INTL. | All Listeria, FDA foods |
| Filter/colony-lift | Carroll et al. (13) | Peer review | L. monocytogenes Meat |
| DNA probe colony hybridization | BAM Chapter 24 (17,18) | FDA | L. monocytogenes, FDA foods |
Alternatively, the methods shown in Table 6 may be used. Identify isolates by conventional or rapid methods. When all Listeria are enumerated estimate the proportion that is L. monocytogenes by determining the species of 10 typical Listeria colonies. For advice on using these techniques consult . Grant (23a) has developed a filter enumeration method, based on the BAM Listeria enrichment and isolation method, which enumerates Listeria at cell numbers of >100 cfu/g.
Tolerance enumeration. Enumeration to determine if a regulated level of tolerance is being met is not needed with the current "zero-tolerance" policy of no detectable L. monocytogenes in 2 x 25-g analytical portions of food or beverage. It would require narrower confidence limits than does surveillance enumeration. Narrower confidence limits for tolerance and surveillance enumeration can be accomplished by counting more colony forming units, which can be accomplished by increasing the number of replicate tubes or other containers. For the current FDA method, the wells of one or more 96-well micro-titer plates, with round-bottomed wells, can be inoculated, by multi-channel pipette or robotically, with 0.1 ml of homogenate of complete BLEB and sample. After incubation at 30° C for 48 h, use the same kinds of transfer methods described in Surveillance Enumeration to inoculate enriched samples to ALOA, BCM or equivalent differential agar to determine which wells are positive. Using the proportion of L. monocytogenes-positive wells, the mean concentration can be calculated using the Poisson equation.
Alternatively, the 1600 filter grid compartments MPN method for Listeria (20) may be used for presumptive enumeration of L. monocytogenes.
Identify isolates by conventional methods, including the use of ALOA, BCM or equivalent agar, or by rapid methods. When necessary estimate the proportion of L. monocytogenes among 10 Listeria isolates.
Simultaneous detection and enumeration. Most samples are likely to be negative and thus it is efficient to delay enumeration of reserve samples until the Listeria detection stage is completed. Even then, most positive samples will only contain a few cfu/25g. Nevertheless it may sometimes be more convenient to do simultaneous detection and enumeration. To accomplish this, prepare the enrichment homogenate as described above and immediately spread 0.1 ml on ALOA, BCM or an equivalent L. monocytogenes selective agar. Incubate plates at 35° C for 24-48 h. The combined minimal method will allow the cell number of presumptive L. monocytogenes to be categorized as <0.04 cfu/g, 0.04 - 100 cfu/g, 100-25,000 cfu/g, or > 25,000 cfu /g. More replica plates and more decimal dilutions in TSBye are optional to obtain a more precise enumeration. Test 5 representative colonies for ability to ferment L-rhamnose by the conventional fermentation method, by the BCM rhamnose confirmatory agar or by a rapid L. monocytogenes identification kit to definitively rule out the uncommon occurrence of L. ivanovii in foods.
References
1. AOAC Official Method 992.18. 2000. MICRO-ID Listeria. Chapter 17.10.02, pp. 141-144 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
2. AOAC Official Method 992.19. 2000. Vitek GPI and GNI+. Chapter 17.10.03, pp. 144-147 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
3. AOAC Official Method 993.09. 2000. Listeria in dairy products, seafoods, and meats. Colorimetric deoxyribonucleic acid hybridization method (GENE-TRAK Listeria Assay). Chapter 17.10.04, pp. 147-150 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
4. AOAC Official Method 993.12. 2000. Listeria monocytogenes in Milk and Dairy Products, Selective Enrichment and Isolation Method (IDF Method). Chapter 17.10.01, pp. 138-139 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
5. AOAC Official Method 994.03. 2000. Listeria monocytogenes in dairy products, seafoods, and meats. Colorimetric monoclonal enzyme-linked immunosorbent assay method (Listeria -Tek). Chapter 17.10.05, pp. 150-152 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
6. AOAC Official Method 995.22. 2000. Listeria in foods. Colorimetric polyclonal enzyme immunoassay screening method (TECRA Listeria Visual Immunoassay [TLVIA]). Chapter 17.10.06, pp. 152-155 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
7. AOAC Official Method 996.14. 2000. Assurance Polyclonal Enzyme Immunoassay Method. Chapter 17.10.07, pp. 155-158 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
8. AOAC Official Method 997.03. 2000. Visual Immunoprecipitate Assay (VIP). Chapter 17.10.08, pp. 158-160 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
9. AOAC Official Method 999.06. 2000. Enzyme Linked Immunofluorescent Assay (ELFA) VIDAS LIS Assay Screening Method. Chapter 17.10.09, pp. 160-163 In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
9a. AOAC Official Method 2003.12. 2003. Evaluation of BAX® Automated System for the Detection of Listeria monocytogenes in Foods. Chapter 17.10.--, pp. --- - --- In: Official Methods of Analysis of AOAC INTERNATIONAL. 17th Edition. W. Horwitz (ed.). Volume 1. Agricultural Chemicals, Contaminants and Drugs. AOAC INTERNATIONAL, Gaithersburg, MD.
10. Alden, M.J., L. Marconi, J. Hogan, I.G. Rosen and R. Johnson. 1990. A chemiluminescent DNA probe assay for the identification of Listeria from culture plates. ICAAC 1990 Abstracts p. 109.
10a. Asperger, H., H. Heistinger, M. Wagner, A. Lehner and E. Brandl. 1999. A contribution of Listeria enrichment methodology - growth of Listeria monocytogenes under varying conditions concerning enrichment broth composition, cheese matrices and competing microflora. Microbiology 16: 419-431.
11. Bille, J., J. Rocourt, and B. Swaminathan. 1999. Listeriae, Erysipelothrix , and Kurthia, pp. 295-314. In: Manual of Clinical Microbiology. 7th Edition. P. R. Murray (ed.). American Society for Microbiology, Washington, DC.
12. Boerlin et al. 1992. L. ivanovii subsp. londoniensis subsp. novi. Int. J. Syst. Bacteriol. 42: 69-73.
13. Carroll, S. A., L. E. Carr, E. T. Mallinson, C. Lamichanne, B. E. Rice D. M. Rollins, and S. W. Joseph. 2000. Development and evaluation of a 24-hour method for the detection and quantification of Listeria monocytogenes in meat products. J. Food Protect. 63: 347-353.
14. Curiale, M. S., T. Sons, L. Fanning, W. Lepper & D. McIver. 1994. Deoxyribonucleic acid hybridization method for the detection of Listeria in dairy products, seafoods, and meats: collaborative study. J. AOAC INTERNATIONAL 77: 602-617.
15. Curiale, M. S., W. Lepper & B. Robison. 1994. Enzyme-linked immunoassay for detection of Listeria monocytogenes in dairy products, seafoods, and meats: collaborative study. J. AOAC INTERNATIONAL 77: 1472-1489.
16. Curtis, G. D. W., R. G. Mitchell, A. F. King, and J. Emma. 1989. A selective differential medium for the isolation of Listeria monocytogenes. Lett. Appl. Microbiol. 8: 95-98.
17. Datta, A. R., B. A. Wentz, D. Shook, and M. W. Trucksess. 1988. Synthetic oligodeoxyribonucleotide probes for detection of Listeria monocytogenes. Appl. Environ. Microbiol. 54: 2933-2937.
18. Datta, A.R., M. A. Moore, B.A. Wentz, J. Lane. 1993. Identification and enumeration of Listeria monocytogenes by non-radioactive DNA probe colony hybridization. Appl. Environ. Microbiol. 59: 144-149.
19. Duvall, R. E., and A. D. Hitchins. 1997. Pooling of non-collaborative multilaboratory data for evaluation of the use of DNA probe test kits in identifying Listeria monocytogenes strains. J. Food Protect. 60: 995-997.
20. Entis, P., and I. Lerner. 2000. Twenty-four-hour direct presumptive enumeration of Listeria monocytogenes in food and environmental samples using the ISO-GRID method with LM-137 agar. J. Food Protection 63: 354-363.
21. Feldsine, P. T., A. H. Lienau, R. L. Forgey, and R. D. Calhoon. 1997. Assurance polyclonal enzyme immunoassay (EIA) for detection of Listeria monocytogenes and related Listeria species in selected foods: collaborative study. J. AOAC INTERNATIONAL 80: 775-790.
22. Feldsine, P. T., A. H. Lienau, R. L. Forgey & R. G. Calhoon. 1997. Visual immunoprecipitate assay (VIP) for Listeria monocytogenes and related Listeria species detection in selected foods: collaborative study. J. AOAC INTERNATIONAL 80: 791-805.
23. Gangar, V., M. S. Curiale, A. D'Onorio, A. Schultz, R. L. Johnson, and V. Atrache. 2000. VIDAS® Enzyme-linked immunofluorescent assay for detection of Listeria in foods: collaborative study. J. AOAC INTERNATIONAL 83: 903-918.
23a. Grant, M. L. Modification of the Bacteriological Analytical Manual Procedure to Permit Enumeration of Listeria monocytogenes by Membrane Filtration. FDA/DFS/ORA Laboratory Information Bulletin 17: (3) LIB No. 4240, 9pp.
24. Higgins, D. L., and B. J. Robison. 1993. Comparison of MICRO-ID Listeria method with conventional biochemical methods for identification of Listeria isolated from food and environmental samples: collaborative study. J. AOAC INTERNATIONAL 76: 831-838.
25. Hitchins, A. D. 1998. Listeria monocytogenes. Chapter 10. In: G. J. Jackson (Coordinator) Bacteriological Analytical Manual. 8th Edition. Revision A. AOAC INTERNATIONAL, Gaithersburg, MD.
26. Hitchins, A. D., and R. E. Duvall. 2000. Feasibility of a defined microflora challenge method for evaluating the efficacy of foodborne Listeria monocytogenes selective enrichments. J. Food Protect. 63:1064-1070.
26b. Hunt, J., K. Jinneman, C. Eklund, J. Wernberg, P. Sado, J. Johnson, R. Richter, S. Torres, E. Ayotte, S. Eliasberg, P. Istafanos, D. Bass, N. Kexel-Calabresa, W. Lin , , and C. Barton. 2002. Inter-Laboratory Validation of a Selective Agar using Chromogenic Substrate for Phosphatidylinositol-Specific Phospholipase C (PI-PLC) Activity to Detect Listeria monocytogenes from Foods. Abstracts Annual General Meeting Amer. Soc. Microbiology. P-43, p. 370.
27. Johansson, T. M-L., R. L. Maijala, and J. A. Hirn. 1995. A less hazardous enrichment broth for Listeria without cycloheximide and acriflavine. J. Food Protection 58:1263-1267.
27a. Johnson, J., K. Jinneman, G. Stelma, B. G. Smith, D. Lye, J. Messer, J. Ulaszek, L. Evsen, S. Gendel, R. W. Bennett and A. D. Hitchins. 2000. Atypical Hemolytic Strain of Listeria Difficult to Speciate by a Battery of Accepted Methods. Poster. AOAC INTERNATIONAL Annual Meeting, Philadelphia, PA.
28. Jones, D., and H.P.R. Seeliger. 1986. International committee on systematic bacteriology. Subcommittee the taxonomy of Listeria. Int. J. Syst. Bacteriol. 36:117-118.
29. Jones, D. 1992. Current classification of the genus Listeria. In: Listeria 1992. Abstracts of ISOPOL XI, Copenhagen, Denmark). p. 7-8.
30. Knight, M. T., M. C. Newman, M. Joseph-Benziger Jr., J. R. Agin, M. Ash, P. Sims, and D. Hughes. 1996. TECRA Listeria Visual Immunoassay [TLVIA] for detection of Listeria in foods: collaborative study. J. AOAC INTERNATIONAL 79: 1083-1094.
31. Lee, W. H., and D. McClain. 1986. Improved L. monocytogenes selective agar. Appl. Environ. Microbiol. 52: 1215-1217.
32. Mattingly, J. A., B. T. Butman, M. C. Plank, and R. J. Durham. 1988. A rapid monoclonal antibody-based ELISA for the detection of Listeria in food products. J. AOAC INTERNATIONAL 71:669-673.
32a. Niederhauser, C., U. Candrian, C. Hofelein, H. P. Jermini, H.P. Buhler, and J. Luthy. 1992. Use of polymerase chain reaction for detection of Listeria monocytogenes in food. Appl. Environ. Microbiol. 58:1564-1568.
32b. Niederhauser, C., C. Hofelein, J. Luthy, U. Kaufmann, H.P. Buhler, and U. Candrian. 1993. Comparison of "Gen-Probe" DNA probe and PCR for detection of Listeria monocytogenes in naturally contaminated soft cheese and semi-soft cheese. Res. Microbiol. 144: 47-54.
33. Restaino, L., E. W. Frampton, R. M. Irbe, G. Schabert, and H. Spitz. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food Protect. 62: 244-251.
34. Rocourt, J., P. Boerlin, F.Grimont, C. Jacquet, and J-C. Piffaretti. 1992. Assignment of Listeria grayi and Listeria murrayi to a single species, Listeria grayi, with a revised description of Listeria grayi. Int. J. Syst. Bacteriol. 42: 171-174.
35. Rocourt, J. 1999. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification. In: Listeria, Listeriosis and Food Safety. E. T. Ryser and E. H. Marth (Eds). 2nd edition, pp. 1-20. Marcel Dekker, Inc., New York, NY.
36. Ryser, E. T., and E. H. Marth. 1999. Listeria, Listeriosis and Food Safety. 2nd edition. Marcel Dekker, Inc., New York, NY.
37. Seeliger, H.P.R., and D. Jones. 1986. Listeria. pp. 1235-1245. In: Bergey's Manual of Systematic Bacteriology, Vol. 2, 9th ed. P.H.A. Sneath, N.S. Mair, M.E. Sharpe, and J.G. Holt (Eds). Williams & Wilkins Co., Baltimore, MD.
38. Sorin, M-L. et al. 2000. Transia Plate Listeria monocytogenes detection in food using an ELISA-based method. IAFP Congress Poster, Atlanta, GA. August.
39. Stelma, G.N., Jr., A. L. Reyes, J. T. Peeler, D.W. Francis, J.M. Hunt, P L. Spaulding, C.H. Johnson, and J. Lovett. 1987. Pathogenicity testing for L. monocytogenes using immunocompromised mice. J. Clin. Microbiol. 25:2085-2089.
40. Twedt, R. M., A. D. Hitchins, and G. A. Prentice. 1994. Determination of the presence of Listeria monocytogenes in milk and dairy products. International Dairy Federation method. J. AOAC INTERNATIONAL 77:395-402.
41. USDA/FSIS. 1999. Isolation and identification of Listeria monocytogenes from red meat, poultry, egg and environmental samples. Ch. 8. Microbiology Laboratory Guidebook. 3rd Edition, Revision 2.
41a. US FDA/CFSAN. 2001. Guidance for Industry Control of Listeria monocytogenes in Refrigerated, Ready-To-Eat Foods. (In preparation)
41b. US DHHS/FDA/CFSAN and USDA/FSIS. 2001. Draft assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods http://www.foodsafety.gov/~dms/lmrisk.html
42. Van Netten et al. 1989. Liquid and solid selective differential media for the detection and enumeration of Listeria monocytogenes. Int. J. Food Microbiol. 8: 299-316.
43. Wang, S-Y. and A. D. Hitchins. 1994. Differential enrichment kinetics of severely and moderately injured Listeria monocytogenes cell fractions of heat injured populations. J. Food Safety 14:259-27.
Assurance EIA Kit and VIP Kit
(Applicable to dairy foods, red meats, pork, poultry products, fruits, nutmeats, seafood, pasta, vegetables, cheese, animal meal, chocolate, and eggs)
Milk, non-fat, dry
Ice cream
Poultry, raw
Shrimp, raw
Beef, cooked roast
Beans, green
GENE-TRAK Listeria Hybridization Assay
(Applicable to dairy products, meats, and seafoods)
Milk, 2%
Cheese, Brie
Crab meat, cooked
Frankfurters
Beef, roast
Pork, raw ground
Listeria -Tek ELISA Assay
(Applicable to dairy products, seafoods, meats)
Frankfurters
Beef, roast, vacuum-packed
Cheese, Brie
Milk, 2%, pasteurized
Shrimp, raw, shelled, frozen
Crab meat, cooked, frozen
TECRA Listeria Visual Immunoassay
(Applicable to dairy foods, seafoods, poultry, meats (not raw ground chuck), leafy vegetables)
Fish, fillets
Ice cream
Lettuce
Chicken
Turkey, ground
VIDAS LIS Assay Screening Method
(Applicable to dairy products, vegetables, seafoods, raw meats and poultry, and processed meats and poultry)
Ice cream
Green beans
Fish
Turkey
Cheese
Roast beef
Abbreviations: P = Pre-collaborative study; C = Collaborative study, "." = No data.
The listed matrices are those encountered by FDA laboratory analysts as of January 1998.
| Food Matrix | Rapid Test Kits | |||||
|---|---|---|---|---|---|---|
| Assurance-EIA and VIP kits | Gene-Trak | ListeriaTek | Tekra VIA | VIDAS Listeria | ||
| Condiments | Mayonnaise | . | . | . | . | . |
| Mustard | . | . | . | . | . | |
| Crustacean | Crab | . | C | . | . | . |
| Crab_cakes | . | . | . | . | . | |
| Crab_ck_fr | . | . | C | . | . | |
| Crab_claws | . | . | . | . | . | |
| Crab_meat | P | P | P | P | P | |
| Crab_meat_cocktail | . | . | . | . | . | |
| Crab_meat_deviled | . | . | . | . | . | |
| Crab_meat_imitation | . | . | . | . | . | |
| Crab_snow (3) | . | . | . | . | . | |
| Crab_stuffed | . | . | . | . | . | |
| Crabmeat | . | . | . | . | . | |
| Crabmeat_backfin | . | . | . | . | . | |
| Crawfish | . | . | . | . | . | |
| Crawfish_chinese | . | . | . | . | . | |
| Crawfish_tails (2) | . | . | . | . | . | |
| Extruded | . | . | . | . | . | |
| Lobster-imitation | . | . | . | . | . | |
| Lobster_meat (4) | . | . | . | . | . | |
| Lobster_meat_cooked | . | . | . | . | . | |
| Lobster_meat_imit. | . | . | . | . | . | |
| Prawns_cooked | . | . | . | . | . | |
| Shrimp | . | . | . | P | P | |
| Shrimp_breaded (2) | . | . | . | . | . | |
| Shrimp_cooked (3) | . | . | . | . | . | |
| Shrimp_peeled | . | . | . | . | . | |
| Shrimp_raw | C | P | C | . | . | |
| Shrimp_salad | . | . | . | . | . | |
| Dairy | Cheese_unnamed (2) | . | . | . | . | CP |
| Cheese_blue | . | . | . | . | . | |
| Cheese_blue_cashel | . | . | . | . | . | |
| Cheese_brie | . | C | C | . | . | |
| Cheese_cheddar | . | P | . | . | . | |
| Cheese_costello | . | . | . | . | . | |
| Cheese_cottage | . | P | . | . | . | |
| Cheese_cows-milk | . | . | . | . | . | |
| Cheese_cream_lox | . | . | . | . | . | |
| Cheese_gorgonzola | . | . | . | . | . | |
| Cheese_ilha_azul | . | . | . | . | . | |
| Cheese_korsholm | . | . | . | . | . | |
| Cheese_kosher-lox | . | . | . | . | . | |
| Cheese_mozzarella | . | . | . | . | . | |
| Cheese_pimento | . | . | . | . | . | |
| Cheese_queso-fr (2) | . | . | . | . | . | |
| Cheese_reblochon | . | . | . | . | . | |
| Cheese_ricotta_hard | . | . | . | . | . | |
| Cheese_ricotta | . | . | . | . | . | |
| Cheese_ricotta_pec | . | . | . | . | . | |
| Cheese_salad-spread | . | . | . | . | . | |
| Cheese_semi_soft | . | . | . | . | . | |
| Cheese_soft | P | . | . | P | . | |
| Cheese_soft_mexican | . | . | . | . | . | |
| Cheese_spread | . | . | . | . | . | |
| Cheese_swiss | . | . | . | . | . | |
| Cheese_white | . | . | . | . | . | |
| Cheese_white_soft | . | . | . | . | . | |
| Cheese_Island | . | . | . | . | . | |
| Cheese_Island_wheel | . | . | . | . | . | |
| Cheese_St.Jorge(2) | . | . | . | . | . | |
| Cheese_St.Jorge_Isl | . | . | . | . | . | |
| Egg_dry | P | . | . | . | . | |
| Egg_liq._frozen | P | . | . | . | . | |
| Ice cream_unnamed | C | P | P | C | C | |
| Ice cream_chocolate | . | . | . | . | . | |
| Ice cream_cookie_flv | . | . | . | . | . | |
| Ice cream_peppermint | . | . | . | . | . | |
| Ice cream_raisin_Fr. | . | . | . | . | . | |
| Ice cream_rum_French | . | . | . | . | . | |
| Ice cream_sandwich | . | . | . | . | . | |
| Ice cream_strawberry | . | . | . | . | . | |
| Ice cream_vanil_crch | . | . | . | . | . | |
| Ice cream_vanl_Fr(2) | . | . | . | . | . | |
| Ice cream_vanilla(2) | . | . | . | . | P | |
| Milk | P | . | P | . | P | |
| Milk_chocolate | P | P | P | P | . | |
| Milk_nf_dry | C | P | P | P | P | |
| Milk_raw | . | . | . | . | . | |
| Milk_2% | . | C | C | . | . | |
| Sherbert_raspberry | . | . | . | . | . | |
| Yogurt_mix_bluebrry | . | . | . | . | . | |
| Yogurt_mix_vanilla | . | . | . | . | . | |
| Fishery | Belt-fish (2) | . | . | . | . | . |
| Calamari | . | . | . | . | . | |
| Catfish (2) | . | . | . | . | P | |
| Catfish_fillet | . | . | . | . | . | |
| Catfish_nuggets | . | . | . | . | . | |
| Chubs, raw | . | . | . | . | . | |
| Chum_roe_salted | . | . | . | . | . | |
| Cuttle-fish_salted | . | . | . | . | . | |
| Cuttlefish | . | . | . | . | . | |
| Cuttlefish_seasoned | . | . | . | . | . | |
| Eel_frozen | . | . | . | . | . | |
| Eel_broiled | . | . | . | . | . | |
| Eel_seasoned | . | . | . | . | . | |
| Eel_smoked | . | . | . | . | . | |
| File-fish_dried | . | . | . | . | . | |
| Fish_barbequed | . | . | . | . | . | |
| Fish_cake | . | . | . | . | . | |
| Fish_cake_fried | . | . | . | . | . | |
| Fish_fillet | . | P | . | C | . | |
| Fish_nova_chips | . | . | . | . | . | |
| Fish_raw | P | . | P | . | CP | |
| Fish_salted | . | . | . | . | . | |
| Fish_smoked (3) | . | . | . | . | . | |
| Fish_stuffed | . | . | . | . | . | |
| Flounder_stuffed | . | . | . | . | . | |
| Flying-fish_roe_sd | . | . | . | . | . | |
| Flying-fish_roe | . | . | . | . | . | |
| Gefilte_fish | . | . | . | . | . | |
| Halibut_fillet | . | . | . | . | . | |
| Halibut_smoked | . | . | . | . | . | |
| Halibut_smoked | . | . | . | . | . | |
| Mackeral_dried | . | . | . | . | . | |
| Pollack_roe (4) | . | . | . | . | . | |
| Pollack_roe_salted | . | . | . | . | . | |
| Pollack_roe_seasnd | . | . | . | . | . | |
| Pollack_roe_spicy | . | . | . | . | . | |
| Orange_roughy_stufd | . | . | . | . | . | |
| Sable_fish_smoked | . | . | . | . | . | |
| Salmon_caviar | . | . | . | . | . | |
| Salmon_jerky | . | . | . | . | . | |
| Salmon_pate | . | . | . | . | . | |
| Salmon_patties (2) | . | . | . | . | . | |
| Salmon_rainbow_smkd | . | . | . | . | . | |
| Salmon_roe_caviar | . | . | . | . | . | |
| Salmon_smoked (5) | . | . | . | . | . | |
| Salmon_smoked_scot | . | . | . | . | . | |
| Salmon_smoked_Atc | . | . | . | . | . | |
| Salmon_sockeye_smok | . | . | . | . | . | |
| Sea-squirt | . | . | . | . | . | |
| Shad_cold-smoked | . | . | . | . | . | |
| Squid_rings | . | . | . | . | . | |
| Sturgeon_smoked | . | . | . | . | . | |
| Surimi | . | . | . | . | . | |
| Swordfish_smoked | . | . | . | . | . | |
| Trout_gldn_rainbow | . | . | . | . | . | |
| Trout_smoked | . | . | . | . | . | |
| Tuna_smoked (2) | . | . | . | . | . | |
| Walleye_fish-fillet | . | . | . | . | . | |
| White-bass | . | . | . | . | . | |
| White-fish_smoked | . | . | . | . | . | |
| Fruit | Cantaloup_sliced | . | . | . | . | . |
| Meal | Bone_meal | P | . | . | . | . |
| Meat | Beef_cooked | . | . | P | . | . |
| Beef_raw | P | . | . | P | P | |
| Beef_roast | C | C | P | P | CP | |
| Franks/Hotdog | . | C | C | P | P | |
| Ham_boiled_slices | . | . | . | . | . | |
| Ham_chopped | . | . | . | . | . | |
| Ham_cured | . | . | P | . | . | |
| Pork_raw | P | C | . | . | . | |
| Pork_raw_ground | . | P | . | . | . | |
| Sausage_bologna | . | . | . | P | P | |
| Sausage_fermented | . | P | . | . | . | |
| Sausage_pork | . | . | . | . | . | |
| Sausage_raw | . | . | P | . | P | |
| Mollusks | Clam_baby_meat | . | . | . | . | . |
| Clams _stuffed (2) | . | . | . | . | . | |
| Clams_baby | . | . | . | . | . | |
| Clams_jack-knife | . | . | . | . | . | |
| Mussells_green_shell | . | . | . | . | . | |
| Mussels_cooked | . | . | . | . | . | |
| Mussels_smoked | . | . | . | . | . | |
| Oysters | . | . | . | P | . | |
| Oysters_fresh | . | . | . | . | . | |
| Oysters_raw | . | . | P | . | . | |
| Scallop_imitat. (2) | . | . | . | . | . | |
| Scallops | P | . | . | . | . | |
| Nuts | Chocolate | P | . | . | . | . |
| Nuts | P | . | . | . | . | |
| Pasta | Pasta_unnamed | P | . | . | . | . |
| Rotini | . | . | . | . | . | |
| Tortellini_cheese | . | . | . | . | . | |
| Poultry | Chicken | . | . | . | P | . |
| Chicken_raw | P | . | P | C | P | |
| Poultry_raw | C | . | . | . | . | |
| Turkey (2) | . | . | . | . | . | |
| Turkey_ground | . | . | . | C | . | |
| Turkey_raw | . | . | P | . | CP | |
| Turkey_raw_ground | . | P | . | . | . | |
| Salads | Cole-slaw | . | . | . | . | . |
| Crab_meat_imitation | . | . | . | . | . | |
| Crab_seafood | . | . | . | . | . | |
| Fish_whiting | . | . | . | . | . | |
| Fruit | . | . | . | . | . | |
| Ham | . | . | . | . | . | |
| Lobster | . | . | . | . | . | |
| Lobster_seafood | . | . | . | . | . | |
| Potato (3) | . | . | . | . | . | |
| Seafood (2) | . | . | . | . | . | |
| Toss_salad | . | . | . | . | . | |
| Sandwiches | Beef | . | . | . | . | . |
| Beef_roast (2) | . | . | . | . | . | |
| Beef_roast_po-boy | . | . | . | . | . | |
| Beef_rst/turkey/chs | . | . | . | . | . | |
| Cheese (2) | . | . | . | . | . | |
| Cheese_burger (2) | . | . | . | . | . | |
| Chicken & swiss | . | . | . | . | . | |
| Chicken_charboiled | . | . | . | . | . | |
| Chicken_salad | . | . | . | . | . | |
| Cold_cut | . | . | . | . | . | |
| Egg_sausage muffin | . | . | . | . | . | |
| Ham & cheese (6) | . | . | . | . | . | |
| Ham & salmon sub. | . | . | . | . | . | |
| Ham & turkey (2) | . | . | . | . | . | |
| Ham/swiss_rye | . | . | . | . | . | |
| Ham_egg_chse_muff. | . | . | . | . | . | |
| Ham_salad | . | . | . | . | . | |
| Hamburger_artific. | . | . | . | . | . | |
| Hamburger_cheese | . | . | . | . | . | |
| Hot_dog_submarine | . | . | . | . | . | |
| Perogie | . | . | . | . | . | |
| Potato/cheese | . | . | . | . | . | |
| Protein_vegetable | . | . | . | . | . | |
| Rib_barbeque | . | . | . | . | . | |
| Steak | . | . | . | . | . | |
| Turkey (4) | . | . | . | . | . | |
| Turkey & cheese | . | . | . | . | . | |
| Turkey_club | . | . | . | . | . | |
| Speciality | Capelin_dried | . | . | . | . | . |
| Chicken_frd_mtless | . | . | . | . | . | |
| Guacamole (2) | . | . | . | . | . | |
| Guacomole_chili | . | . | . | . | . | |
| Tofu | . | . | . | . | . | |
| Vegetables | Avocado_pulp (2) | . | . | . | . | . |
| Beans_green | C | . | . | . | C | |
| Blend_frozen | . | . | . | . | . | |
| Broccoli_frozen | . | . | . | . | . | |
| Broccoli_normandy | . | . | . | . | . | |
| Cabbage_shredded | . | . | . | . | . | |
| Carrots_crinkle-cut | . | . | . | . | . | |
| Coleslaw | P | . | . | . | . | |
| Lettuce | . | . | . | C | . | |
| Mixed_rotini | . | . | . | . | . | |
| Salad_mix_veggies | . | . | . | . | . | |
| Salad_veggies_shred | . | . | . | . | . | |
| Spinach | . | . | . | . | . | |
Hypertext Source: Bacteriological Analytical Manual, 8th Edition, Revision A, 1998. Chapter 10.