Intramural Targeted Anti-COVID-19 (ITAC) Awards
In response to the COVID-19 crisis, the Office of Intramural Research organized the Intramural Targeted Anti-COVID-19 (ITAC) funding program. With the financial support of NIAID, the program provided $12 million to intramural investigators for research into understanding and/or combatting this dangerous disease. The program provided funding for 40 of the 159 applications received. These funded projects are described here.
Early Cell-Free DNA Profiles to Predict COVID-19 Clinical Trajectories
Awardee
Sean Agbor-Enoh, M.D., Ph.D. (NHLBI)
Abstract
At COVID-19 onset, patients generally show mild symptoms. Thereafter, their clinical trajectories vary significantly ranging from minimal symptoms only to multisystemic disease manifesting with different combinations and severities of non-specific symptoms, cytokine storm, respiratory failure, thromboembolic disease, and others. These trajectories also vary by race. Unfortunately, there are no early predictors. Further, the sources and trends of tissue injury contributing to these diverse phenotypes remain poorly defined. This study leverages the high sensitivity of plasma cell-free DNA (cfDNA) and cell-free mitochondrial DNA (mtcfDNA), biomarkers of tissue injury that are also damage-associated molecular patterns (DAMPs). cfDNA is an early predictive biomarker. In transplantation for example, cfDNA detects rejection 2 – 3 months earlier than biopsy. Further, early post-transplant cfDNA levels vary between patients and predict subsequent cfDNA trends and long-term outcomes. Therefore, we hypothesize that cfDNA is an early predictive marker of COVID-19 clinical trajectories and outcomes. To address this hypothesis, we propose a multicenter prospective cohort study to recruit COVID-19 patients, collect serial plasma samples, perform cfDNA measures, and test if cfDNA predict COVID-19 clinical trajectories and outcomes. In exploratory studies, we will test if cfDNA and mtcfDNA contribute to COVID-19 pathogenesis as DAMPs.
Sneaking Out with the Trash: How Coronaviruses Exit Cells
Awardee
Nihal Altan-Bonnet, Ph.D. (NHLBI)
Abstract
Beta-Coronaviruses are enveloped positive strand RNA viruses that include major human and animal pathogens such as Middle East Respiratory Syndrome Virus (MERS) and the Severe Acute Respiratory Syndrome Viruses SARS CoV1 and SARS CoV2. Beta-Coronaviruses cause major host morbidity and mortality and by targeting the cells of the pulmonary, cardio, circulatory and immune systems, and induce complex disease phenotypes. In particular the host innate and adaptive immune systems appear to be dysregulated by these viruses with significant negative consequences on the host such as uncontrolled inflammation and cytokine storms. What the fundamental underlying cause for this immune dysregulation is currently unknown. Contrary to existing dogma, our studies have revealed that Beta-Coronaviruses use lysosomes to exit and spread to other cells (Ghosh et al., submitted 2020) and this profoundy disrupts lysosome physiology with lysosomes being deacidified and lysosomal proteolytic enzymes being inactivated. We hypothesize that this major disruption of normal lysosomal physiology will profoundly disrupt the normal regulation of the host innate and adaptive immune systems as lysosomes are critical for antigen presentation; for cytotoxic cell killing; and for phagocytic degradation of pathogens. This proposal aims to understand the impact of viral disruption of lysosome physiology on the host immune system; and identify viral and host machinery regulating viral lysosomal exit for targeted antiviral development.
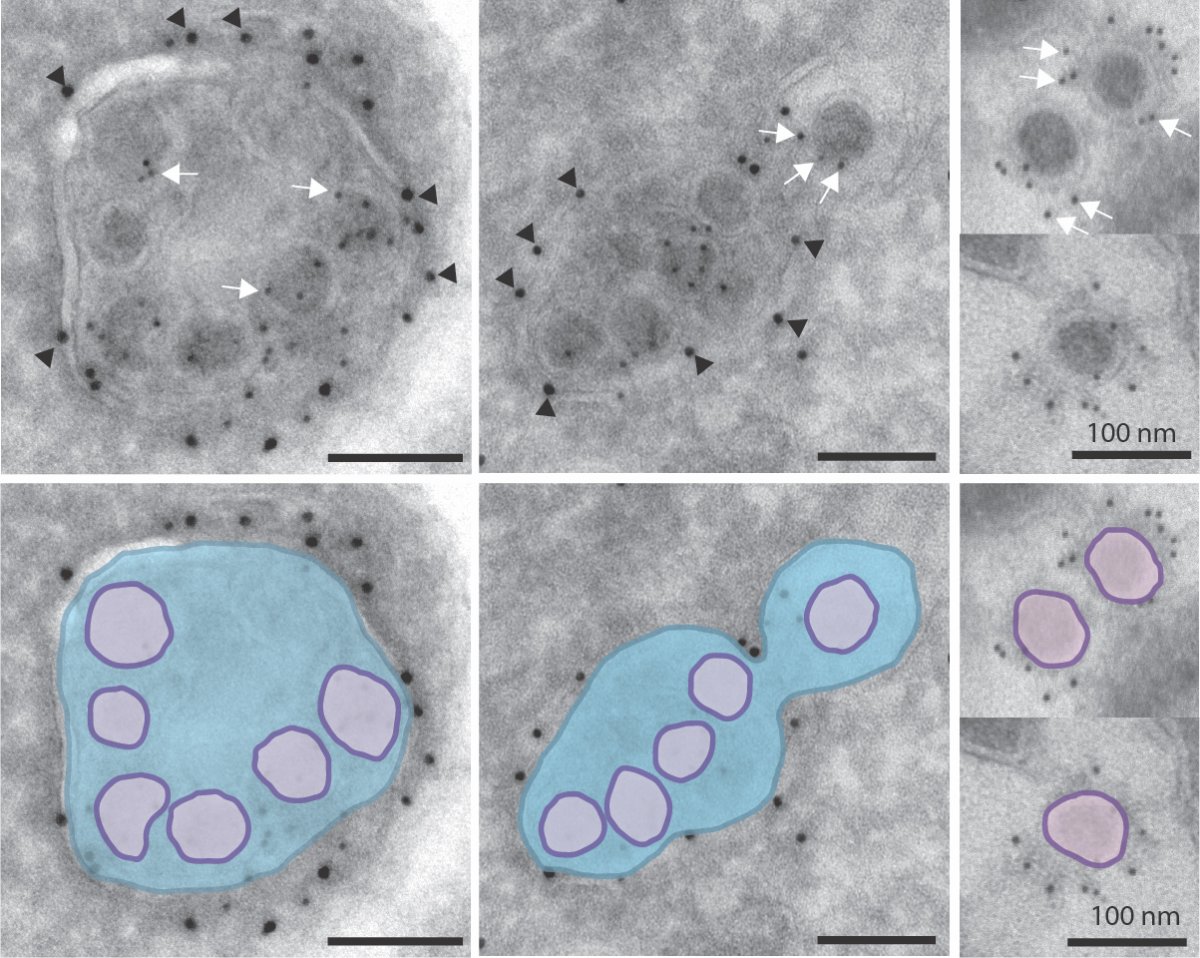
Coronaviruses inside lysosomes imaged by EM.
Novel Lung Imaging Methods to Evaluate Patients with COVID-19
Awardee
Adrienne Campbell, Ph.D. (NHLBI)
Abstract
Imaging technologies developed at DIR/NHLBI offer a unique opportunity to meet the urgent need for improved diagnosis and monitoring of COVID-19, and for understanding of the history and mechanisms of the disease. These include ultra-low dose CT technology that permit repeated examination at short intervals, MRI and CT technologies that provide ultra-detailed information on tissue structure and function. In addition to COVID-19 patients at NIH, we propose a collaboration with the INOVA Fairfax hospital and Medstar Washington Hospital Center that treat a large number of COVID-19 patients to develop, validate, and use these technologies to study COVID-19.

Adeno Associated Virus Based Therapeutics Against SARS-CoV-2
Awardee
Jay (John) Chiorini, Ph.D. (NIDCR)
Abstract
Emerging viruses such as SARS-CoV-2 represent a significant risk to the global security and public health. Although vaccine technology has advanced, the development of new approaches to combat viral infections are needed. In recent years, the use of broadly neutralizing antibodies or nanobodies to control virus infection has shown promise and even resulted in approved drugs. However, limitations to using these therapeutic vaccine technologies is the short half-life of these antibodies in vivo and the need for repeated injections of the neutralizing antibodies. Over the past 10 years, gene delivery technology based on adeno associated virus (AAV) has made rapid progress and has generated approved drugs for treating genetic diseases. The AAV gene delivery platform offers the possibility of employing a single application for potential life-long expression of a therapeutic protein. In this proposal we plan to develop and utilize AAV based delivery of biologically engineered proteins to test both the immune stimulatory potential as well as the expression of SARS-CoV-2 neutralizing nanobodies or antibodies. By investigating different AAV vectors in the specific context of developing antibody-based immunity, we can provide insight into both novel candidate platforms for the delivery of new antiviral agents as well as test new therapeutics modalities for blocking the spread of the virus within a community.
Deciphering the Double-Edged Role of IFITM3 During SARS-CoV-2 Infection
Awardee
Alex Compton, Ph.D. (NCI)
Abstract
Interferon-induced transmembrane protein 3 (IFITM3) is a critical component of the cell-intrinsic immune response to respiratory pathogens such as Influenza A virus. However, its role in controlling coronaviruses is unclear. In cell culture, IFITM3 has been shown to inhibit SARS-CoV-1 but it promotes infections by other human coronaviruses. Furthermore, mutations in IFITM3 toggle its antiviral versus proviral effects. It was recently reported that a single nucleotide polymorphism in IFITM3 is associated with COVID19 disease severity. Here, we propose to address the impact of IFITM3 on recently-emerged SARS-CoV-2. In addition to the receptor used by the virus for cellular attachment, ACE2, it was shown that SARS-CoV-2 can utilize a host cell protease known as TMPRSS2 to cleave and activate viral spike protein for fusogenic activity. In our preliminary experiments utilizing HIV-based pseudovirions bearing SARS-2-CoV spike, we found that IFITM3 does not inhibit, but rather promotes, infection in cells lacking TMPRSS2. In contrast, IFITM3 expression in TMPRSS2-positive cells results in potent inhibition of infection. Using cell culture models and in vivo models of infection, the goal of this proposal is to understand the mechanistic interplay between TMPRSS2, IFITM3, and SARS-CoV-2 entry into cells. The results of our aims will enable a better understanding of SARS-CoV-2 tropism in vivo and its propensity for disease.
Epitope Dynamics of Native SARS-CoV-2 S Protein by Cryo-Electron Tomography
Awardee
William Copeland, Ph.D. (NIEHS)
Abstract
Corona viruses get their name from the spikes exposed on the membrane surface. This fusogenic S-protein constitutes a major viral antigen. Since it is required to recognize and enter target cells, mechanisms have evolved that reduce its immunological footprint while preserving its function. This is achieved through a complex set of conformational changes combined with extensive glycosylation. Understanding the structural dynamics of the SARS-CoV-2 S-protein is crucial for the rational design of vaccines and neutralizing agents. S protein is a trimer anchored to the viral envelope by a transmembrane domain. The native spike is metastable and detergent solubilization can trigger conformational changes leading to misfolded protein. Stabilized surrogates lacking the transmembrane domain have been engineered for serological tests and as vaccine candidates, and their structure solved using cryo-electron microscopy (cryo-EM). While facilitating structural studies of S protein in complex with various agents, this approach has serious limitations. Subtle differences in the sequence of spike variants have been linked to significant changes in antigenicity. It has also been shown that the neutralization of envelope glycoproteins by antibodies targeting the membrane proximal region often involve interactions with lipids. Further, alterations in the cytoplasmic domain are suspect of triggering conformational changes in the ectodomain. Therefore, the structural identity of the truncated ectodomain with the native spike cannot be guaranteed. We propose to deploy our high throughput workflows in cryo-EM of viral particles and macromolecules to elucidate the in-situ structure and conformational landscape of native S-protein and its complexes with neutralizing antibodies and drugs.
Control of Ribosomal Frameshifting on the SARS-CoV-2 mRNA
Awardee
Thomas Dever, Ph.D. (NICHD)
Abstract
Viral replication is dependent on the host protein synthesis machinery, and many viruses exploit fidelity controls of host translation to modulate expression of key viral proteins. Translation of the gag-pol precursor protein of SARS-CoV-2 is dependent on programmed -1 ribosomal frameshifting between the gag and pol open reading frames (ORFs). A fraction of ribosomes translating the gag ORF, shift reading frames in a -1 direction at a specific ‘slippery’ sequence to synthesize the gag-pol fusion protein. Altering the frequency of frameshifting and thus the ratio of gag to gag-pol proteins impairs replication of a variety of viruses including SARS-CoV-1, raising the possibility that targeting frameshifting might be of therapeutic value in treating COVID-19. Recently, the interferon-induced gene C19orf66 encoding Shiftless was reported to impair -1 ribosomal frameshifting on the HIV genome. We propose to test whether Shiftless impacts frameshifting during translation of the SARS-CoV-2 mRNA. Relatedly, in unpublished work, we found that altering the function of translation factors eIF5A and eEF2 affects translational frameshifting, and we propose to test the role of these factors in SARS-CoV-2 frameshifting. We will use yeast, mammalian cell, and in vitro reconstitution studies to examine the impacts of Shiftless, eEF2, and eIF5A on SARS-CoV-2 frameshifting. Small molecule modulators of eIF5A function are already known and will be tested for impacts on SARS-CoV-2 frameshifting. These studies and the assays developed in this proposal could be used to screen for small molecules that affect frameshifting and that might be of potential therapeutic value in COVID-19.
Clonal Hematopoiesis and Severe COVID-19 Disease in Humans and Macaques
Awardee
Cynthia Dunbar, M.D. (NHLBI)
Abstract
COVID-19 disease caused by infection with the novel SARS-CoV-2 coronavirus is extremely heterogenous, ranging from asymptomatic infection to a flu-like illness all the way to respiratory failure and widespread additional organ damage. Profound inflammation is the hallmark of severe COVID-19, and generally does not occur until the 2nd week of infection, when viral replication may already be decreasing. While risk factors for this late hyperinflammatory syndrome have been identified, most notably age greater than 60 and/or a number of pre-existing conditions such as diabetes, hypertension, and cardiopulmonary disease, in many cases specific factors resulting in severe COVID-19 disease remain elusive. Some elderly patients have mild disease and conversely younger patients without identified risk factors can develop hyperinflammatory disease. Rhesus macaques have been rapidly established as a model for SARS-CoV infection and are being utilized to test therapies and vaccines. However, macaques do not develop late hyperinflammatory disease, and thus cannot be used to investigate the pathophysiology of this outcome or test therapeutic approaches. We hypothesize that one explanation for the increased incidence of severe COVID-19 disease with age as well as the heterogeneity of progression could be the impact of acquired somatic mutations in certain genes in hematopoietic stem and progenitor cells (HSPC), termed age-related clonal hematopoiesis (ARCH). Loss of function mutations in several epigenetic regulators, most commonly TET2 and DNMT3A, have been linked to clonal expansion of HSPC and a marked hyperinflammatory phenotype, specifically involving IL6 and IL1? pathways. ARCH begins to increase markedly beginning in the 6th decade, matching the time frame for increased COVID-19 morbidity and mortality. We propose to perform targeted sequencing of ARCH genes in two cohorts of patients with COVID-19 disease and age-matched controls, asking whether patients with severe COVID and hyperinflammatory markers have a higher incidence of ARCH. If so, individuals with ARCH and COVID could be targeted for early interventions interrupting implicated inflammatory mediators. We have developed a robust rhesus macaque model for ARCH via CRISPR/Cas9 editing of the TET2 locus in young adult HSPC followed by autologous transplantation, reproducing the clonal expansion and hyperinflammatory phenotype observed in older humans with ARCH. We propose to use this model to ask whether animals with engineered ARCH develop late severe disease following SARS-CoV-2 infection, in contrast to control macaques. If so, this will represent an invaluable model to understand the pathophysiology of severe disease, as well as allowing insights into the link between ARCH, hyperinflammation and COVID disease outcomes via intensive immune cell phenotyping, single cell RNASeq and analysis of cytokines and other inflammatory mediators.
Taste Receptor Cells Harbor SARS-CoV-2
Awardee
Josephine Egan, M.D. (NIA)
Abstract
SARS-CoV-2 is a coronavirus that is the cause of the present global pandemic. The ACE2 enzyme is its receptor, while TMPRSS2 facilitates viral infection in mammalian cells. Tissue localization of the receptor and protease correlates with presentation of symptoms, and organ dysfunction and infection are more virulent in cells that express ACE2 on cilia. Continued viral shedding in asymptomatic and pre-symptomatic individuals enhances community transmission. Chemosensory disruption is a symptom of SARS-CoV-2 infection. Taste receptor cells (TRCs) in taste buds embedded in taste papillae in the tongue express both ACE2 and TMPRSS2 and TRCs have cilia. Human taste buds are an immune-privileged site as they lack surveilling immune cells. Our hypothesis is that human TRCs possess both the machinery for, and provide an ideal environment for, SARS-CoV-2 infection and are a site for asymptomatic infection that is emerging as a significant problem in containing community spread. Serendipitously, we have an active, IRB-approved protocol to obtain human fungiform papillae to study TRC and taste stem cell function with age and in age-related diseases. We have a biorepository of cells from over 100 papillae taken from people aged 22-80 years of age, some with pre-existing conditions e.g. diabetes and cancer survivors. We have cultured and propagated taste organoids from these biopsies and are poised to pivot and begin work to again propagate organoids so as to infect them with SARS-CoV-2. We can next investigate the potential of many classes of compounds to impede SARS-CoV-2 infection and proliferation in our human taste organoids.
Impact of SARS-CoV2 Infection in Pulmonary Vascular Endothelium
Awardee
Jason Elinoff, M.D. (NIH Clinical Center)
Abstract
While hypoxic respiratory failure requiring mechanical ventilation is the predominant clinical manifestation of critical illness with severe acute respiratory syndrome coronavirus-2 (SARS-CoV2), pulmonary vascular and cardiovascular manifestations including pulmonary embolism and shock are among the most frequent causes of death. Yet, a mechanistic understanding of these vascular complications is lacking. Our laboratory specializes in cellular models of endothelial dysfunction in the context of pulmonary arterial hypertension, a severe, chronic pulmonary vascular disease characterized by a proliferative, pro-thrombotic and inflammatory cellular phenotype. Here we plan to investigate the cellular, molecular and transcriptomic consequences of SARS-CoV2 infection of human primary endothelial cells in clinically relevant vascular compartments (e.g. pulmonary artery, pulmonary microvascular, cardiac microvascular, coronary artery, systemic artery and vein). Access to blood samples and clinical phenotypes of a cohort of subjects with severe COVID-19 infection admitted to the NIH Clinical Center ICU will facilitate in vivo translation of in vitro mechanistic insights.

Development and characterization of endothelial cell models of SARS-CoV2 infection.
Structural Elucidation of the SARS-Cov-2 RNA Genome for Antiviral Discovery
Awardee
Adrian Ferré-D'Amaré, Ph.D. (NHLBI)
Abstract
The coronaviruses have the largest genomes of all RNA viruses. The 5' and 3' untranslated regions (UTRs) of these viruses have highly conserved sequences that have been proposed to adopt complex three-dimensional structures. The high conservation of these elements, including in SARS-CoV-2, suggests that these elements perform critical functions. Within the 5' UTR, stem-loops 2 and 3 (SL2, SL3) contain the transcriptional regulatory sequences necessary for discontinuous synthesis of subgenomic negative-strand RNA. Also within the 5' UTR, SL5 contains the start codon of ORF1ab, which signals the beginning of the coding region, and is involved with viral RNA packaging. The 3' UTR contains the mutually exclusive bulged stem-loop/pseudoknot region, which may control viral replication in a switch-like fashion. The structures of these elements remain unknown. Since compounds that target complex RNA folds have a long history of successful development into therapeutics (for instance, more than 80% of antibiotics in clinical use target the bacterial ribosomal RNA), and we have recently been successful in structure-guided discovery of high affinity ligands for even relatively small (~30 kilodalton) structured RNAs, we propose to characterize these elements biochemically and biophysically, and to determine their structures at atomistic resolution. Being less than 80 kilodaltons in size, none of these elements are suitable for cryo-electron microscopic analysis. Thus, we plan to determine their structures by X-ray crystallography. Success of work in this proposal will set the stage for subsequent structure-guided drug discovery efforts.
Development of 3D Cellular Lung Models for Antiviral Testing
Awardee
Marc Ferrer, Ph.D. (NCATS)
Abstract
The NCATS 3D Tissue Bioprinting Laboratory (3DTBL) exists as a core resource to collaborate with the scientific community to biofabricate functional human tissues in multi-well format to enable disease modeling, predictive toxicology, and pre-clinical drug testing. High-impact respiratory viruses pose a significant threat to global health. The ongoing COVID-19 pandemic, caused by the recently emerged SARS-CoV2, highlights the need for well-characterized, physiologically-relevant cellular systems for respiratory viral disease modeling and drug discovery. To address this need, the NCATS 3DTBL is developing 3D lung tissue equivalents produced with human primary lung epithelial cells, pericytes, and vascular endothelial cells. These in vitro lung tissue equivalents will be used to generate pathophysiologically relevant infection models for human respiratory viruses, including seasonal (e.g. HCoV-NL63, RSV, PIV, influenza) and newly emerging (e.g. SARS-CoV2, ) respiratory viral infections. The 3D lung tissue platform will be established and validated using BSL2 respiratory viruses and with disease-relevant assay readouts such as viral infectivity, cytokine secretion, fibrosis, and edema. Furthermore, because specific determinants and detailed host-cell interactions that make a respiratory virus capable of causing severe disease remain unknown, we propose to use single-cell RNA sequencing to identify host transcriptomic responses to respiratory viral infection and disease progression. By thoroughly establishing the robustness of this translatable pan-viral human 3D lung-respiratory assay platform using BSL2 respiratory viruses, we will ensure that it is also amenable for rapid deployment to investigate BSL3 viral threats like SARS-CoV2, enabling the understanding of disease progression and as a tool for drug discovery.

Investigation of 25-Hydroxycholesterol as a Therapeutic for COVID-19
Awardee
Michael Fessler, M.D. (NIEHS)
Abstract
The recent rapid international spread of SARS-CoV-2 is a global health emergency. To date, there is no proven therapy for the associated infectious disease, COVID-19. Recently, 25-hydroxycholesterol (25HC), a native oxysterol generated by the action of the enzyme cholesterol-25-hydroxylase upon cholesterol, has been shown to be broadly antiviral. 25HC has potently inhibited every enveloped virus tested to date, including Coronaviridae, and is thought to act by blocking virus-cell fusion. Systemic treatment of mice with 25HC antagonizes viral infections in vivo and one study showed that 25HC was therapeutic in pigs against respiratory infection with highly pathogenic PRRSV, a virus within the same order (Nodoviridae) as coronaviruses. 25HC was moreover shown to be effective against Zika virus in rhesus macaques with no evident toxicity during a 7-day course, suggesting it may be safe in humans. Our recently published work has shown that 25HC is also anti-inflammatory in the mouse lung. We hypothesize that 25HC antagonizes SARS-CoV-2 infection by inhibiting virus-cell fusion, and that 25HC will be therapeutic against SARS-CoV-2 respiratory infection. To address this hypothesis, we propose the following two Specific Aims: (i) Determine whether 25HC inhibits SARS-CoV-2 infection of respiratory epithelial cells; and (ii) Determine whether 25HC is therapeutic in vivo against SARS-CoV-2 respiratory infection. Our proposal employs several innovative tools, including human lung organoids, enantiomeric 25HC, mouse-adapted virus, and pseudotyped reporter virions for assay of viral entry and fusion. Given the preclinical safety profile of 25HC, confirmation of efficacy against SARS-CoV-2 could lead to its rapid testing in humans.
Antagonism of Coronavirus Spike Proteins by Cellular Host Factors
Awardee
Eric Freed, Ph.D. (NCI)
Abstract
The coronavirus (CoV) spike (S) proteins play an essential role in viral entry by binding the cell- surface receptors on target cells and mediating the fusion between viral and cellular membranes during virus entry. The S protein is also the target of neutralizing antibodies generated by the infected host. Because of its central role in virus infection and adaptive immunity, the S protein is a prime target for the development of antiviral therapeutics and vaccines. Viral infections trigger an innate immune response, largely induced by interferon (IFN), that sets up an antiviral state in infected cells and tissues. Hundreds of interferon-stimulated genes (ISGs) are induced by viral infection. While the role of specific ISGs in blocking the replication of particular viruses has been well established, the vast majority of ISGs have not been characterized. Because of the significance of host innate immunity in viral transmission and replication within and between hosts, there is an unmet need to understand these antiviral inhibitory factors in detail. We plan to apply our many years of experience studying the biogenesis, trafficking, and function of HIV envelope (Env) proteins to address key questions related to the role of host factors in antagonizing CoV S proteins. In this proposal, we outline a focused and high-impact series of experiments aimed at elucidating the role of two families of ISGs – the membrane-associated RING CH domain (MARCH) E3 ubiquitin ligases and the guanylate binding proteins – in antagonizing the S proteins of SARS-CoV, SARS-CoV-2, and MERS-CoV.

The MARCH8 E3 ubiquitin ligase antagonizes the spike (S) protein of SARS-C0V-2. On the left is shown the topology of MARCH8, with the RING E3 ubiquitin ligase domain indicated. On the right, western blot analysis of virus particle-associated S protein in the absence of MARCH8, or in the presence of WT or catalytically inactive MARCH8 in the virus producer cell. Levels of S protein precursor and the cleaved product S2 are severely reduced by MARCH8 expression.
Imaging Effect of S-Protein Mediated ACE2 Downregulation on Lung Injury
Awardee
Dima Hammoud, M.D. (NIH Clinical Center)
Abstract
Angiotensin-converting enzyme 2 (ACE2) was recently identified as a functional receptor for Severe Acute Respiratory Syndrome coronavirus-2 (SARS-CoV-2) entry into human host cells, similar to what was previously described for SARS-CoV. The majority of ACE2 expressing cells in the lungs are alveolar epithelial type II (AECII) cells [1], which apparently have multiple viral process related genes. This explains the early involvement of the lungs with the infectious process, with AECII facilitating viral replication. ACE2, however, also has a lung protective role that has been shown using animal models of lung injury such as acid aspiration, cecal ligation/perforation, and intratracheal LPS administration in ACE2 knockout mice. Interestingly, recombinant human ACE2 (rhACE2) decreased the degree of lung injury in ACE2 knockout mice in response to acid administration, further supporting the protective role of ACE2 in the lungs. In the setting of SARS-CoV-2 infection, the viral spike (S) protein is assumed to bind to ACE2, resulting in internalization and destruction of the receptor. We hypothesize that in Covid-19, SARS-CoV-2 S-protein-mediated ACE2 downregulation contributes to the severity of lung pathology, not only because of direct infection, but also because of the dysregulation of ACE2 expression, an important lung protective pathway. We are proposing an in vivo imaging study with small animal high-resolution CT and FDG PET imaging in human-ACE2 transgenic mice (K18-hACE2), wild type mice and ACE2 Knockout mice to demonstrate the effects of ACE2 downregulation on lung injury, in response to S-protein exposure. To induce acute lung injury (ALI) in our mouse model, we will use intranasal Lipopolysaccharide (LPS). We will corroborate our imaging findings with immunostaining and quantitative measures of lung injury and ACE2 expression in our animal models. Finally, we will use rhACE2 to evaluate its potential for rescuing hACE2 mice from the effects of spike protein administration and preventing worsening of the lung injury.
Targeting ER Proteostasis to Improve Outcome from SARS-CoV-2 Infection
Awardee
Brandon Harvey, Ph.D. (NIDA)
Abstract
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a readily-transmitted, novel viral pathogen belonging to the Coronoaviradae family and is responsible for causing the current COVID-19 pandemic. The SARS-CoV-2 virus was first associated with respiratory-related symptoms in humans, buy it is now known to affect multiple tissue systems. Like many viruses, SARS-CoV-2 exploits the functions of the endoplasmic reticulum (ER) within their host cells to produce viral proteins and lipid membranes. Several viral proteins from the highly homologous SARS-CoV-1 localize to the ER and Golgi of a host cell and form functional ion channels in the membrane known as “viroporins.” The SARS-CoV-1 viral proteins E, 3a and 8a have been identified as viroporins, and both E and 3a are essential for viral replication and virulence. The E protein causes calcium ion permeability in the ER/Golgi membrane. One of the primary functions of the ER is to store and buffer calcium, and high calcium concentration in the ER is important for maintaining ER proteostasis and retention of ER resident proteins. SARS-CoV-1 viroporins decrease ER calcium. Our lab recently discovered that the ER proteome is redistributed following depletion of ER calcium, specifically, proteins containing an ER retention sequence are secreted from the cell. This mass departure of ER resident proteins, referred to as “exodosis” represents a potential pathogenic mechanism that can be countered by stabilizing ER calcium. The consequences of exodosis are both a loss of protein function from the ER (e.g. chaperone, isomerase, peptidase) and gain of function outside the cell. We previously developed a luciferase-based reporter (GLuc-SERCaMP) that can monitor the exodosis phenomenon. In collaboration with NCATS, we used the GLuc-SERCaMP reporter to develop a target agnostic screen for drugs that stabilize ER calcium and prevent exodosis. We screened over 100,000 compounds and identified several FDA-approved drugs and novel drugs that can stabilize ER calcium and the ER proteome. In this proposal, we will examine 1) the effects of SARS-CoV-2 viroporin proteins (E, 3a, 8a) on ER calcium homeostasis and ER proteostasis in human cell lines from different tissue origins, 2) the effects of FDA-approved drugs that were identified as modulators of ER calcium for their ability to modulate ER stress associated with expression of SARS-CoV viroporins, 3) the effects of top FDA-approved and novel drugs identified as modulators of ER calcium for their ability to alter the unfolded protein response, inflammatory response, and cell viability following SARS-CoV-2 infection of human lung epithelial cells. Successful completion of the project would provide new insight into the biology of SARS-CoV-2 related to the function of the endoplasmic reticulum in host cells and identify a therapeutic approach to reduce ER-associated cellular toxicity and inflammation caused by SARS-CoV-2 infection.
Development of Neutralizing Nanobodies Against SARS-CoV-2
Awardee
Mitchell Ho, Ph.D. (NCI)
Abstract
COVID-19 has dealt a devastating blow to global health and the worldwide economy. Neutralizing antibodies against SARS-CoV-2 and other related coronaviruses can be useful in treating this ongoing pandemic. In this proposal, we plan to use our unique single domain (also called “nanobody”) phage display libraries derived from camels and sharks to identify novel cross-neutralizing nanobodies against both SARS-CoV-2 and SARS-CoV, and potentially MERS-CoV as well. Due to their small size and high stability, nanobodies might have the ability to be administered by an inhaler making them attractive therapeutics for respiratory infections. Our research plan aims to (i) screen nanobodies to SARS-CoV-2 spike (S) protein using our large single domain phage libraries and to (ii) analyze the biological and structural features of nanobodies. SARS-CoV-2 enters into a human cell through its S protein binding to angiotensin-converting enzyme 2 (ACE2). Therefore, the receptor binding domain (RBD) of the S protein is the primary target for neutralizing antibodies. Our pilot screen has already identified a panel of camel nanobodies that might recognize diverse conformations of the RBD. Some might bind an “open” conformation of the RBD, which might block the interaction with the ACE2 on human cell, while others might bind a “close” conformation, which might lock its conformation in the off state. We envision that a cocktail strategy combining two (or more) nanobodies that recognize different parts including neutralizing and non-neutralizing epitopes of the viral surface that interact with human cells might be the most effective in the future.

Isolation of neutralizing nanobodies targeting SARS-CoV-2. In the receptor binding stage, the S1 subunit of SARS-CoV-2 binds human ACE2 on the host cell surface. Nanobodies bind the RBD domain on the S1 subunit might block the interaction of the RBD and the ACE2. In the viral fusion stage, after the cleavage of S1 subunit, the viral fusion peptide (FP) on the S2 subunit inserts into the host cell membrane, inducing the conformational change of the S2 subunit, which forms a six-helix bundle (6-HB) with the HR1 and HR2 trimers. Nanobodies that target the HR domains might block viral fusion.
Development of Cell-Based Assays to Identify SARS-CoV-2 Protease Inhibitor
Awardee
Wei-Shau Hu, Ph.D. (NCI)
Abstract
SARS-CoV-2 relies on two viral proteases to cleave and activate its RNA genome replication machinery. These two proteases, papain-like (PL) and 3-chymotrypsin-like (3CL, also known as the main) protease, are excellent targets for inhibitors because their functions are essential for viral replication. Indeed, a promising experimental intervention for COVID-19 uses a 4-drug combination that includes two repurposed HIV-1 protease inhibitors, lopinavir and ritonavir, which inhibit the 3CL protease. The PL and 3CL proteases are highly conserved between SARS-CoV and SARS-CoV-2, and these two proteases have been targets for SARS-CoV inhibitor development. However, there are very few mammalian cell-based assays to screen for 3CL protease inhibitors. Most existing assays use purified proteins or replication-competent viruses. Purified protein-based assays are sensitive to assay conditions and do not account for cellular absorption or toxicity of compounds. Propagation of replication-competent virus requires a BSL-3 containment laboratory, which is not conducive to high-throughput screening. In this application, we propose to develop human cell-based assays to identify inhibitors of 3CL proteases in BSL-2 containment. We have previously developed a FRET-based assay to monitor activity of HIV-1 protease. We will adapt this assay to monitor the activity of the SARS-CoV-2 3CL protease. As an alternative approach, we will develop two bimolecular complementation-based assays to monitor inhibitor activity. After developing the assays, we will screen a library containing FDA-approved drugs to identify compounds that have the potential to be repurposed for COVID-19 treatment. Additionally, we will extend these assays to identify broad-spectrum inhibitors of human coronaviruses.
Regulation of Thrombo-Inflammatory Mechanisms in COVID-19
Awardee
Yogendra Kanthi, M.D. (NHLBI)
Abstract
There are increasing reports of arterial and venous thrombosis in patients with coronavirus disease 19 (COVID-19). Dysregulation of tonic vascular homeostatic functions of endothelial and hematopoietic lineage cells has been implicated in the pathophysiology of COVID-19. Biomarker studies and lung histopathology have revealed a marked imbalance of coagulation and fibrinolysis, but the underlying mechanisms remain elusive. Understanding the mediators of endothelial and platelet dysfunction that provoke unimpeded thrombin generation, and defining the role of prothrombin-targeting antibodies will provide a critical foundation to identify more precise and effective therapeutic opportunities to target the thrombo-inflammatory coagulation cascade in COVID-19.
Modulation of Chronic Inflammatory Responses by COVID-19
Awardee
Mariana Kaplan, M.D. (NIAMS)
Abstract
The COVID19 pandemic is revealing pleiotropic clinical manifestations and outcomes that are, in many cases, driven by the inflammatory response that ensues. The role of genetic and environmental factors in shaping the immune response to this infection remains to be characterized. Understanding how individuals with chronically dysregulated immune systems, such as adult and pediatric patients with systemic autoimmune diseases, respond to COVID-19 exposure, and how this infection and the ensuing immune response modulates the immune dysregulation and organ damage characteristic of these diseases, may not only provide crucial information to physicians treating patients with these conditions, but will also improve the scientific knowledge of mechanisms or immune-mediated injury, tissue damage and vasculopathy from this infection in the general population. We have established a systemic autoimmunity/COVID19 research group encompassing 3 NIH institutes. Given that a significant proportion of the population will be exposed to COVID-19 over the next couple of years, it will be important to: 1) characterize how COVID-19 modulates systemic inflammation, autoimmunity features, organ damage and vasculopathy in adult and pediatric patients with a previous diagnosis of systemic autoimmunity, as well as its potential regulation by demographic factors, immune-modulators, antivirals and other therapies; 2) assess how subjects with systemic autoimmunity respond to COVID-19 infection regarding antiviral and/or proinflammatory responses , response to potential treatments and vaccine, and overall outcomes. These aims will also contribute to understand prevalence and severity of this infection in individuals with autoimmune diseases, and the variables that associate/predict these responses. Subjects with various systemic autoimmune diseases will be assessed prior to infection and post-infection to establish these outcomes through clinical evaluation as well as by assessing genetic determinants, transcriptomics (bulk and single cell both oil peripheral blood and available tissues), autoantibody repertoire modulation, innate and adaptive immune cell subsets, cytokine responses and vascular function and arterial wall inflammation. Overall, the results of this study will significantly contribute to understanding the modulation of inflammatory and vasculopathic responses in subjects with dysregulated immune systems, following exposure to a new type of viral challenge. These results will contribute not only to better understand how the virus is impacting people with chronic inflammatory conditions but may also allow the identification of new therapeutic targets for the acute and chronic complications of this potentially devastating viral illness.
Genetic Susceptibility to Severe Respiratory Disease Induced by SARS-CoV2
Awardee
Steven Kleeberger, Ph.D. (NIEHS)
Abstract
Respiratory infectious diseases resulting from bacterial or viral pathogens such as Mycobacterium tuberculosis, Streptococcus pneumoniae, respiratory syncytial virus (RSV), and influenza, are major global public health concerns. Lower respiratory tract infections are leading causes of morbidity and mortality, behind only ischemic heart disease and stroke. The morbidity and mortality associated with the current COVID-19 pandemic caused by SARS-CoV-2 threaten to supersede those of all other respiratory diseases. Since late January, the virus has infected over 4.1 million people globally and, while a majority of infected people recover, over 284,000 have died (W.H.O.). As with other viral infections, some human subpopulations are particularly vulnerable. Many factors may contribute to the inter-individual variation in response to respiratory infections, including gender, age, socioeconomic status, nutrition, pre-existing conditions or diseases (e.g. respiratory disorders, diabetes, obesity, cardiovascular disorders), and genetic background. Little is known about the genetic determinants of susceptibility to COVID-19, though gene candidates have been proposed. In collaboration with colleagues in Brazil and Argentina, we have considerable experience in identification of genetic variants that confer susceptibility to severe RSV disease in infected infants. We have also developed a genetic diagnostic single nucleotide polymorphism (SNP) panel to identify individuals that are highly susceptible to severe RSV disease. We propose herein to 1) identify SNP variants in SARS-CoV-2 infected individuals that predispose to severe disease caused by the virus and 2) develop a diagnostic SNP panel that may be used to screen otherwise healthy individuals who may be highly susceptible to SARS-CoV-2-induced disease.
Discovery and Development of Novel Antivirals Against SARS-CoV-2
Awardee
T. Jake Liang, M.D. (NIDDK)
Abstract
In as short as 4 months, COVID-19 has spread and ravaged the world in an unprecedented speed in modern history rivaling the 1918 flu pandemic. SARS-CoV-2, the culprit virus, is highly contagious and stable in the environment and predominantly transmits among humans via the respiratory route. The virus does not only wreak havoc in the lung, but also inflict systemic diseases involving the gastrointestinal tract, cardiovascular system, kidney, and neurological system. Currently, no clinically approved therapies or vaccines are available for this disease, with the exception of remdesivir for severely ill patients with Covid-19. The overall goal of this proposal is to identify and develop effective antivirals against the SARS-CoV-2, either by repurposing existing pharmaceuticals or developing new drugs. Here we plan to establish non-infectious cell-based model systems to study various stages of SARS-CoV-2 infection and replication cycle, to develop high-throughput platform based on these model systems to screen large small-molecule libraries for anti-SARS-CoV-2 compounds, and to conduct extensive preclinical studies of highly active and nontoxic compounds from the screen for further drug development.
The COVID-19 Pandemic Vulnerability Index
Awardee
Alison Motsinger-Reif, Ph.D. (NIEHS)
Abstract
Synthesis of diverse and disparate data is crucial for decision-makers, particularly at the state and local levels, to respond to the COVID-19 pandemic. To facilitate state and local official’s needs to effectively prioritize resources, identify and address key vulnerabilities, and evaluate and implement effective interventions during the COVID-19 Pandemic, we developed the COVID-19 Pandemic Vulnerability Index (PVI) Dashboard and accompanying county-level Scorecards that integrates key data and shows which indicators contribute to local vulnerability (https://covid19pvi.niehs.nih.gov/). Individual data streams comprised of well-established general vulnerability factors for public health disasters and factors emerging as relevant to the COVID-19 pandemic have been aggregated and displayed in a graphical arcGIS context. The Dashboard visualizes vulnerability indicators in a radar chart, where each of the indicators comprises a “slice” of an overall PVI profile. This PVI highly associated with local case and death outcomes. The Dashboard is updated daily with county-level data and supports interactive evaluation of current data for all U.S. counties in an overall view, plus detailed county-level PVI Scorecards. The data visualization in this dashboard offers an effective means of communicating data to scientists, to policy makers, and to the public. Currently, we propose to continue to add emerging, relevant data streams, and to add robust statistical predictions for forecasting. Additionally, we will harden the web-based tool and build additional software tools to empower further modeling. While numerous Dashboards are emerging related to COVID19, ours is unique in the large number of data sources assembled and the clear visualization of local conditions.
Single-Molecule Measurement of COVID-19 Replication Inhibition
Awardee
Keir Neuman, Ph.D. (NHLBI)
Abstract
Severe respiratory syndrome related coronavirus-2 (COVID-19) is the most recent severe coronavirus to emerge as a threat to human health on a global scale. Understanding and refining current therapies and finding new ways to combat viral infection and prevent viral amplification are critical to not only mitigate the current outbreak but also prepare for future viral pandemics. One promising antiviral target is the replication/transcription complex (RTC) that is responsible for both amplifying genomic RNA and production of viral mRNA. Two key core RTC enzymes are the RNA dependent RNA polymerase (RdRP) and the viral helicase that are both highly conserved in sequence and function in contrast to viral structural proteins. The recent success of Remdesivir to treat COVID-19 patients highlights this fact and warrants better understanding of not only how these and other inhibitors work against the viral enzymes, but also how the target proteins function, at the molecular level. In this proposal, we will employ a versatile single molecule technique, magnetic tweezers, to study how the two key replicative proteins, RdRP and helicase function alone and in conjunction, which will establish a baseline understanding of their catalytic mechanisms. This baseline understanding of their activity will permit testing known and novel inhibitors including Remdesivir, which is the main focus of the proposal. We will elucidate how known inhibitors modulate the catalytic function and determine the mechanism, specificity, and effectiveness of these inhibitors at the molecular level in conjunction with molecular dynamic simulations that will facilitate interpretation of the single-molecule measurements.
COVID-19 Pandemic Impact on Alcohol and Related Outcomes
Awardee
Vijay Ramchandani, Ph.D. (NIAAA)
Abstract
The COVID-19 pandemic, resulting from an outbreak of a novel coronavirus (SARS-CoV-2) infection, has become an unprecedented global threat to individuals, communities and health systems. While immediate attention has appropriately focused on prevention and treatment of SARS CoV-2 infection, the widespread societal mental health consequences of the pandemic cannot be ignored. Given the catastrophic impact of the COVID-19 pandemic, it is critical to prospectively and longitudinally assess the impact on alcohol use and problems, along with associated behaviors and outcomes. The goal of this study is to examine the impact of the COVID-19 pandemic on alcohol use and consequences in individuals across the spectrum of alcohol use and alcohol use disorder (AUD). The specific aims of this study are: (1) to evaluate the impact of the COVID-19 pandemic on alcohol consumption and consequences in individuals across the spectrum of alcohol use; (2) to evaluate the time-course of changes in measures of negative life events, social isolation and stress, and their effect on alcohol consumption and consequences during and following the COVID-19 pandemic; (3) to explore the role of anxiety, depression, craving, binge drinking, impaired control in the relationships examined above. The target sample size for this study will consist of 700 participants including, non-drinkers, light drinkers, non-treatment-seeking heavy drinkers, and treatment-seeking individuals with AUD who have been screened under the NIAAA Screening Protocol, thus leveraging the extensive screening and deep phenotyping data obtained from that evaluation. Participants will be invited to complete several surveys by phone and/or online over 2 years at intervals that range from weekly to bimonthly in the first year and every 6 months during the second year. Depending on the trajectory of the pandemic, the frequency of the surveys may be modified if necessary. The surveys will assess a range of outcomes related to alcohol consumption and consequences, along with measures of other substance use, stress, sleep, pain, physical health and quality of life. Study will be conducted via telephone and online survey, and participants will not have in-person study visits. The proposed study includes a prospective longitudinal assessment of the impact of the pandemic on alcohol consumption, consequences, impaired control, stress, sleep and associated measures and outcomes, in individuals with and without AUD prior to the pandemic. This study will provide unique insights into the short- and long-term impact of the pandemic on alcohol use and related outcomes, with the potential to inform future efforts in the prevention and treatment of AUD in individuals exposed to major public health disasters and other stresses, including social isolation, job loss, serious illnesses and deaths in the community, that are salient in the current pandemic.
Pathogenesis and Modulation of Coronavirus-Induced Inflammatory Disease
Awardee
Barbara Rehermann, M.D. (NIDDK)
Abstract
We have established mouse models with natural microbiota (bacteria, viruses, fungi from wild mice) which we plan to use to investigate coronavirus-induced inflammatory disease pathogenesis. WildR mice have pathogen-free natural microbiota that effectively mitigate inflammation, resulting in a 75% decrease of fatality after respiratory virus (flu) infection compared to conventional mice with lab mouse microbiome. Wildling mice with natural microbiota at all body sites and pathogens predict hyperinflammatory immune responses of humans in phase I studies, whereas conventional laboratory mice do not.

This project studies the role of microbes in age-dependent, organ-specific inflammation after coronavirus infection.
Amphipathic Peptides as Antiviral Agents for SARS-CoV-2
Awardee
Alan Remaley, M.D., Ph.D. (NHLBI)
Abstract
SARS-CoV-2, the causative agent of the COVID-19 pandemic, is an RNA-enveloped virus. Disrupting lipid membranes of enveloped viruses with detergents, such as soaps, readily inactivates viruses. Amphipathic peptides can also disrupt lipid membranes, but unlike chemical disinfectants they can be tailored to be more selective, with limited cytotoxicity. Several amphipathic peptides, including those based on apolipoprotein A-I that remove cholesterol from cells, have anti-viral effects on a wide variety of enveloped viruses. By detailed structure-function studies over 20-years, we have developed several apolipoprotein A-I mimetic peptides, one of which is now undergoing a Phase I clinical trial. Using three prototypical amino acids (Glu, Leu and Arg) plus Ala, we will synthesize a large library of peptides with different biophysical properties (charge, size, hydrophobicity, hydrophobic moment) and hypothesize that we will be able to identify an amphipathic peptide that will inactivate SARS-CoV-2. This will be determined by the infection of Vero cells and primary human bronchial epithelial cells by a VSV-pseudovirus expressing the SARS-CoV-2 spike-protein and a model betacoronavirus. We will also investigate the antimicrobial properties of the peptides on bacteria that cause secondary bacterial pneumonia and their cytotoxicity profile on multiple human cell lines. Peptides with a favorable therapeutic index(IC50/CC50) will be tested for virus neutralization of the intact SARS-CoV-2 virus in a BSL-3 cell culture facility. This study will provide insights into the structural and biophysical features necessary to develop novel peptide based therapies for SARS-CoV-2 that could also be effective for other known or emerging pathogenic enveloped viruses.
Epigenetic and Environmental Factors of ACE2 and TMPRSS2 Co-Expression
Awardee
Joseph Rodriguez, Ph.D. (NIEHS)
Abstract
Over the past 4 months the severe acute respiratory syndrome coronavirus SARS-CoV-2 (COVID19) has caused a global pandemic, leading to hundreds of thousands of deaths. Particularly enigmatic is the variability of phenotypes and severity of symptoms exhibited by COVID19 patients. A major area of research is the mode of viral entry into human cells. COVID19 entry requires the expression of the human genes ACE2 and TMPRSS2. ACE2 is the angiotensin-converting enzyme 2 and is used by COVID2 to dock and enter host cells. TMPRSS2 is an androgen and estrogen responsive serine protease which is required for S protein priming of coronaviral spike protein. Interestingly, ACE2 and TMPRSS2 co-expression in the same cells is rare and highly variable in epithelial cells of several tissues. Therefore, in order to understand COVID19 infectivity and susceptibility, a deeper mechanistic understanding of their co-expression is necessary. We propose to identify epigenetic and environmental factors that modulate ACE2 and TMPRSS2 regulation. We hypothesize that ACE2 and TMPRSS coactivation is modulated in part by extrinsic factors such as cell cycle dependent estrogen receptor regulation, and intrinsic factors such as epigenetic regulation at single loci. We propose a high-throughput imaging screen to identify epigenetic and environmental factors that modulate ACE2 and TMPRSS2 co-expression. We will also determine the temporal cell cycle dynamics of ACE2 and TMPRSS2 expression by single molecule transcriptional activity in living cells. This work will help gain a mechanistic view of ACE2 and TMPRSS2 gene activation and the environmental factors that alter their expression.
Targeting Functional RNA Elements in the SARS-CoV-2 Genome
Awardee
John Schneekloth, Ph.D. (NCI)
Abstract
The SARS-Cov-2 virus is a pathogen that has caused remarkable economic damage and mortality in a short period of time. Unfortunately to date there are no drugs that are clinically efficacious against the virus, and new strategies are needed. While most drug discovery approaches aim at inhibiting viral proteins, this proposal takes a different approach: developing small molecule inhibitors that target viral RNAs. The SARS-CoV-2 genome contains multiple functional elements such as the frameshifting element (FSE) and Stem-loop II-like motif (s2m) sequences. These elements exhibit high levels of conservation and are highly structured, making them excellent targets for small molecules. Here, we take advantage of unique infrastructure and expertise of the laboratories of John Schneekloth, Jr. and Stuart Le Grice, both at the National Cancer Institute. We will use a high throughput microarray screening platform to identify druglike chemicals that bind to FSE and s2m RNAs. We will also utilize structural, chemical, and biophysical techniques to develop potent, selective inhibitors of these viral RNAs. This is conceptually novel approach that is needed to develop compounds to be used as tools to study viral infectivity, or to be developed as potential antiviral therapeutics.
Epigenetic Control of ACE2 by the Chromatin Remodeler, SMCHD1
Awardee
Natalie Shaw, M.D., MMSc. (NIEHS)
Abstract
SARS-Cov-2 gains entry into human cells via the ACE2 receptor and also causes a dramatic decrease in ACE2 cell surface expression upon infection, which wreaks havoc on the many organs that rely on a finely tuned balance between angiotensin II and angiotensin(1-7) activities. Thus, it is critical to understand how ACE2 expression is modulated by SARS-Cov-2. Recent studies in mouse heart cells demonstrated mechanical stress-induced recruitment of Brg1, a component of the SWI/SNF chromatin remodeling complex, to the Ace2 promoter, where it repressed Ace2 expression. Following ablation of another chromatin remodeler, SMCHD1, we observed 15-fold upregulation of ACE2 mRNA in human cranial placode cells, which are precursors of olfactory neurons and their support cells. Taken together, these data demonstrate that ACE2 is an epigenetic target and raise the intriguing possibility that SARS-Cov-2 represses ACE2 expression in target tissues by hijacking the host epigenetic machinery. In the proposed studies, we will investigate how SMCHD1 represses ACE2 expression in human cells and will leverage our structural biology program to determine if SMCHD1 itself or one of its protein binding partners may be druggable targets.
Cancer and COVID-19 Surveillance Linkage Study
Awardee
Meredith Shiels, Ph.D. (NCI)
Abstract
SARS-CoV-2 infection may be of particular concern among patients actively being treated for cancer as well as cancer survivors, given that cancer treatment is associated with immune suppression and infection risk. There is an urgent need to quantify COVID-19 risk, severity and mortality among cancer patients and survivors and identify risk factors for poor outcomes. The proposed study will link US state cancer registries and COVID-19 registries to rapidly answer several critical questions for cancer patients and survivors. Specifically, this study will 1) compare risk of severe COVID-19 (i.e., hospitalization, intensive care unit [ICU] admission, and mechanical ventilation) and death across cancer sites; 2) identify risk factors for severe COVID-19 and death among cancer patients and survivors; 3) estimate risk of severe COVID-19 and death among cancer patients and cancer survivors relative to the general population; and 4) estimate the risk of death among cancer patients and survivors with COVID-19 compared to those with COVID-19, but no prior cancer. This registry linkage will be carried out in two phases in June and December 2020. To date, New Jersey, Connecticut and Kentucky have agreed to participate (163,500 cases of COVID-19 and 10,700 deaths) in the first round of linkages, and additional states have inquired about joining later this year. This study will provide much needed information that can be used to understand which cancer patients and survivors are at highest risk of severe COVID-19 and those that have the poorest outcomes, which will inform recommendations for these cancer populations.
Structural and Functional Characterization of SARS-CoV-2 Nsp15
Awardee
Robin Stanley, Ph.D. (NIEHS)
Abstract
SARS-CoV-2 is a single-stranded RNA coronavirus that causes Covid 19 and has led to an unprecedented world-wide health pandemic. There is an urgent need to develop new therapeutics to target this virus. Coronaviruses have a large RNA genome that encodes for 16 non-structural proteins (Nsp) that are important for RNA synthesis and processing. In order to develop new therapeutics to target Covid 19 we need to define the structure, function, and regulation of the Nsp RNA processing factors. Nsp15 is an endoribonuclease that specifically cleaves 3’ of uridines and is well conserved across coronavirus family members. Nsp15 was recently shown to block activation of host viral sensors by specifically cleaving poly-uridines found at the 5’ end of the negative sense RNA strand. Since the RNA target of Nsp15 was just discovered, how Nsp15 recognizes and cleaves poly(U) sequences is poorly understood. This proposal seeks to use a structural and functional approach to define the molecular mechanisms of RNA processing by Nsp15. We recently determined the cryo-EM structure of Nsp15 to atomic resolution and identified conformational dynamics within the nuclease domain. Our central hypothesis is that conformational dynamics of the nuclease domain are critical for RNA processing. Beyond defining the mechanisms of RNA processing we also propose to determine how known inhibitors block Nsp15 activity and how other factors regulate Nsp15 function. Collectively, this work will define the molecular basis for evasion of the host viral response by Nsp15 and reveal how Nsp15 nuclease activity is modulated by inhibitors and co-factors.
Cardiopulmonary Inflammation and Multi-System Imaging in COVID-19 Infection
Awardee
Anthony Suffredini, M.D. (NIH Clinical Center)
Abstract
There is little sequential, methodically collected data on patients with COVID-19 that can be correlated with the severity of disease or the long-term sequelae. Such information is necessary to understand how to most logically intervene with directly acting antiviral agents and immunologic response modifiers. This protocol will enroll patients to the Clinical Center in a longitudinal study (NIH 20-CC-0113). The first cohort (n=36) will be comprised of acutely diagnosed persons with a spectrum of illness i.e. limited symptoms and organ dysfunction to persons requiring hospitalization at the CC. They will be followed acutely, during recovery (at 4 – 8 weeks) and convalescence (8 weeks to 12 months). A second cohort (n=36), recruited from regional medical centers after recovery from COVID-19 infection will similarly be studied during recovery and convalescence. The goal is to link a) inflammatory responses present in blood, bronchoalveolar lavage, urine and spinal fluid with b) viral RNA levels in blood and body fluids and c) innovative imaging of COVID-19 target organs (lungs, heart, brain and kidneys) from the earliest stages of infection and at later time points as the infection and host responses evolve, through recovery and convalescence. We believe that this approach will provide novel insights into mechanisms associated with the initiation, progression and resolution of lung, cardiac and systemic inflammation. These mechanisms are presumed to be essential in the pathogenesis and survival from this infection. This information will help guide diagnostic and therapeutic innovation and assess long-term consequences of this infection.
Structure of SARS-CoV-2 N Protein/RNA Assemblies from CryoEM and ssNMR
Awardee
Robert Tycko, Ph.D. (NIDDK)
Abstract
We propose to characterize the molecular structure of ribonucleoprotein (RNP) assemblies formed by the SARS CoV-2 nucleocapsid (N) protein with RNA. Within mature SARS CoV-2, the viral RNA is associated with multiple copies of N, most likely in a helical assembly. Nearly all structural details of coronavirus RNP assemblies are currently unclear, with only speculative models in the existing literature. We will form homogenous RNP assemblies using recombinant, full-length N protein and use cryo-electron microscopy (cryoEM) to develop a density map for these assemblies. Combined with known structures of the isolated RNA-binding and dimerization domains of coronavirus N proteins, this density map will reveal the arrangement and relative orientations of the folded domains of N in the context of RNP assemblies, the location of RNA, and the nature of protein-protein and protein-RNA interfaces. Additionally, solid state nuclear magnetic resonance (ssNMR) measurements will be performed on isotopically labeled RNP assemblies, as well as microcrystalline states of the RNA-binding and dimerization domains. The ssNMR data will provide information about amplitudes and timescales of molecular motion within the RNP assemblies, especially for segments of N that are unstructured in its monomeric state. Measurements of interatomic distances and torsion angles by ssNMR will complement the cryoEM density map, leading to a high-resolution picture of critical intermolecular interfaces. Finally, time-resolved ssNMR methods developed in our lab will be used to characterize the sequence of events that lead to fully-formed RNP assemblies, starting with unassembled N protein and RNA. Results from this work will contribute to our understanding of the viral assembly process, and will provide a basis for the development of antiviral drugs that target RNP assemblies.
Chimeric ACE2 Peptide Ligand for Diagnostic Assays of SARS-CoV-2
Awardee
Alexander Wlodawer, Ph.D. (NCI)
Abstract
This proposal is to design and examine the functionality of a chimeric peptide comprising three fragments of cell surface receptor ACE2, together forming the epitope recognized by the receptor binding domain (RBD) of SARS-CoV2 S1 spike protein. Potential applications for this construct as a highly specific, physiologically relevant ligand include ultra-sensitive S1 spike antigen detection and quantitation assays using Immuno-qPCR technique applicable to the saliva and mucosal samples and in vivo bioimaging, biodistribution, clearance, and toxicity studies. Development of the testing methods proposed here constitutes a novel approach to detection and assessment of viremia and distribution of the virus throughout body compartments, as well as immune responses in symptomatic and convalescent patients, as well as in asymptomatic carriers. This chimera is designed as a ligand complementary to the SARS-CoV2 S1, based on a highly conserved interaction interface between S1 RBD and ACE2. The design is based on several available high-resolution structures of the complexes. As such it may also represent a promising tool applicable to isolation of diverse viral strains and cross-reactive immunogen design. The novel approaches proposed here are designed to improve the understanding of development of an adaptive immune response to the virus, as a result of either virus exposure or an active infection, as well as to advance coronavirus epidemiology and pathogenicity studies.
Artificial Intelligence from Chest CT to Assess COVID-19 Clinical Trials
Awardee
Bradford Wood, M.D. (NIH Clinical Center)
Abstract
COVID-19 has characteristic features on CT. CT image processing and deep learning models provide quantifiable metrics to serve as a noninvasive biomarker for pulmonary involvement. Correlation with a variety of clinically relevant metadata may enable the use of CT AI during outbreaks to identify CT biomarker features for clinical trials in COVID-19. Artificial intelligence (AI) tools based upon deep learning of large multi-national chest CT datasets provide an opportunity for classification from other pneumonias, as well as for measuring outcomes and response in clinical trials. CT AI models derived from large data sets may define CT phenotypes of disease dynamics, inform immediate isolation of asymptomatic contacts, facilitate drug discovery, clarify chronic lung effects, and perhaps inform or refine outcomes in specific cohorts of clinical trials. A multidisciplinary multi-IC, public-private partnership developed and independently tested and validated CT AI segmentation, classification, and infiltrate quantification models, which could serve as standardized clinical trial response criteria. Public models have been posted for no-cost use, as well as web-based models for academic and developer use, with direct CT upload functionality, and an output of % likely COVID-19 infiltrate present. Such standardized imaging biomarker tools could expedite drug discovery or enhance our understanding of failure modes or response signals. Goals: Facilitate validation of a standardized tool for establishment of public deep learning models for quantification and standard response criteria metrics for characterization of COVID-19 clinical trials and correlation of immune and inflammatory molecular dynamics with CT AI imaging profiles. Hypothesis: CT imaging data aggregation and artificial intelligence will inform and expedite clinical and preclinical studies of COVID-19. SPECIFIC AIMS / EXPERIMENTAL PLAN: COVID-19 clinical trials call for standardized tools for measuring outcomes and response. CT AI models derived from large data sets may define CT phenotypes of disease dynamics, infectiousness, resource allocation and facilitate drug discovery and expedite assessment of therapeutic success in clinical trials.

Chest CT scans and Gradient-weighted Class Activation Maps for a deep learning model for COVID-19 classification. Similar artificial Intelligence algorithms yield automated quantification which might help define response in clinical trials for medical countermeasures or to phenotype failures. The saliency heat maps define the areas used for the decision-making by analyzing the gradient information flowing into the last convolutional layer of the neural network. Reference: https://www.nature.com/articles/s41467-020-17971-2
Visualizing SARS-CoV-2 Virus Entry and Its Underlying Endocytic Mechanisms
Awardee
Ling-Gang Wu, M.D., Ph.D. (NINDS)
Abstract
CoVid-19 infection caused by SARS-CoV-2 virus (CoV2) has become a global pandemic. Studies showed that CoV2, an enveloped, single-stranded RNA virus, enters the cell via endocytosis. However, the underlying endocytic mechanisms, which are crucial for CoV2 entry and clinical prevention of CoV2 entry, remain unclear, owing to difficulty of visualizing CoV2 entry in real time. Recently, we have successfully visualized membrane transition from flat-shape through lambda-shape invagination (inverse V shape) and omega-shape structure to O-shape vesicle during vesicle endocytosis in live secretory cells. With this technical breakthrough, we found that endocytic proteins, such as the cytoskeletal F-actin and the GTPase dynamin, mediate these membrane structural transitions by providing mechanical forces to pull and constrict membrane. Accordingly, I hypothesize that CoV2 enters the cell via endocytic membrane transition from flat-shape through lambda- and omega-shape to O-shape, and that different endocytic proteins, including FCHO, actin, clathrin and dynamin, generate different forces to drive flat membrane through lambda- and omega-shape to O-shape vesicle containing CoV2. We will test this hypothesis by 1) visualizing endocytic membrane dynamics that mediate CoV2 entry with the super-resolution STED microscopy in real time, and 2) determining the mechanical and molecular mechanisms underlying each membrane structural transition that mediates CoV2 entry. The results will uncover how endocytic proteins provide forces to mediate CoV2 entry in real time. The mechanisms discovered here will account for not only CoV2 entry, but also many other viruses (including influenza virus) that enter cells via endocytosis. These findings will conceptually advance our understanding of viral entry, which will ultimately help prevention and treatment of CoV2 infection as well as many other virus infections.
Mechanism of Inhibition of Entry Inhibitors Against SARS-CoVs
Awardee
Di Xia, Ph.D. (NCI)
Abstract
Coronavirus entry into host cells is mediated by the transmembrane spike glycoprotein (S protein) that forms homotrimers protruding from the viral surface. The S protein comprises two functional subunits responsible for binding to the host cell receptor (S1 subunit) and fusion of the viral and cellular membranes (S2 subunit). Thus, S protein is a major vaccine candidate and antiviral target. Using an in vitro cell-based membrane fusion assay, we have successfully identified several small molecular lead compounds as well as virus-derived peptides that inhibit the process of membrane fusion. To further characterize modes of action of these compounds, we propose to determine structures of the S protein or its receptor-binding fragments in complex with the lead compounds in the presence or absence of the host cell receptor ACE-2. We expect that these inhibitory compounds may exhibit different modes of action from destabilizing the prefusion complex to interfering with the formation of the complex between S protein and ACE-2 receptor.
Structural Basis for Inhibiting the COVID-19 RTC
Awardee
Wei Yang, Ph.D. (NIDDK)
Abstract
The novel coronavirus (SARS-CoV-2 (1), which causes Covid-19) has infected more than 4 million people and caused more than 280,000 deaths worldwide (www.worldometers.info/coronavirus) in five months. The pandemic has led to global lockdown and trillions of dollars of financial losses. Without a vaccine or drugs to treat Covid-19, we are still in the lockdown and cannot resume normal daily activities. Furthermore, there is danger of a second wave of the Covid-19 pandemic this coming winter. Coronaviruses are single-stranded RNA virions, and their infections require the virus-encoded RNA-dependent RNA polymerase (RdRp) and helicase for the RNA genome replication. The drugs against Covid-19, Remdesivir and Favipiravir, both target RdRp and have shown promising results in patients with severe symptoms (2). There is no inhibitor of the viral helicase in clinical trial so far. Although structures of RdRp of SARS-CoV-2 and the helicases of SARS and MERS have been reported (3-5), how these enzymes bind the RNA substrate, where most inhibitors work, remains unknown. Building on the successful experience of my group in determining structures of DNA polymerases and helicases in complex with nucleic acid substrates, and in discerning how HIV reverse transcriptase has two binding modes its RNA/DNA substrate, one for the DNA polymerase and one for its RNase H activity, we propose to elucidate high-resolution structures of SARS-CoV-2 RdRp, helicase and accessary proteins in complex with the viral RNA using cryoEM. Our goal is to improve the specificity and potency of RdRp inhibitors and to discover new drug targets.
Parvovirus Particles to Present Coronavirus Spike Antigens in a Vaccine
Awardee
Neal Young, M.D. (NHLBI)
Abstract
We propose chimeric parvovirus capsids to present SARS-Cov-2 antigens in a vaccine. Parvovirus capsid proteins self-assemble to empty capsids. The unique region of the minor capsid protein (VP1u) is external to the capsid and presents most linear neutralizing epitopes to the immune system. In humans, humoral immunity to parvovirus B19 is robust and lifelong. A VP1u-enriched capsid is NIH-sponsored development to prevent parvovirus infection. VP1u can be replaced by heterologous sequences of about 200 amino acids, which retain conformation, remain functional, and elicit immune responses in animals. Capsid proteins are expressed efficiently in engineered yeast. We will design plasmids and express chimeric capsids in yeast, so that parvovirus particles contain SARS-Cov-2 peptides in place of the VP1u spike, including especially 1) the coronavirus receptor binding domain (RBD); or 2) the N-terminus of nucleocapsid protein (N); or 3) a repeated neutralizing epitope of RBD amino acids 485-517, as well as controls (VP2-only, and an irrelevant viral antigen). Capsids would be characterized for content of heterologous peptides and surface localization. Antibodies would be elicited in mice immunized with capsids at two doses with MF59 as adjuvant, and neutralization assessed and quantified in susceptible cell lines in vitro. If a neutralizing humoral response is achieved, nonhuman primates would be challenged after immunization with the optimal capsid preparation and dose. Antigen presentation on a parvovirus platform has the advantages of non-infectious, immunogenic particles, derived from a common benign virus; already piloted in humans; and scalable in a current anti-B19 parvovirus vaccine contract.
The page was last updated on Wednesday, October 14, 2020 - 11:29am




