Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Contact with Dairy Bull Calves
Posted November 28, 2016 12:30 PM ET
Highlights
- Read the Advice to Livestock Handlers and Veterinarians »
- Read Information for Health Care Providers »
- CDC, several states, and the U.S. Department of Agriculture’s Animal and Plant Health Inspection Service (USDA-APHIS) are investigating a multistate outbreak of multidrug-resistant Salmonella Heidelberg infections.
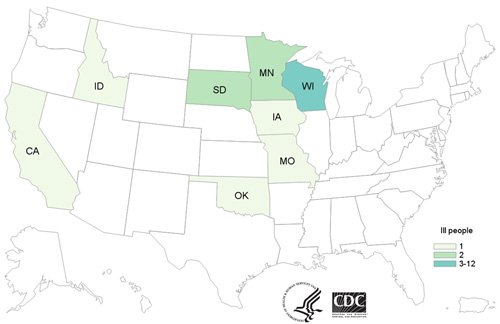
- Twenty-one people infected with the outbreak strain of Salmonella Heidelberg have been reported from 8 states.
- Illness onset dates range from January 11, 2016 to October 24, 2016.
- Eight ill people were hospitalized, and no deaths have been reported.
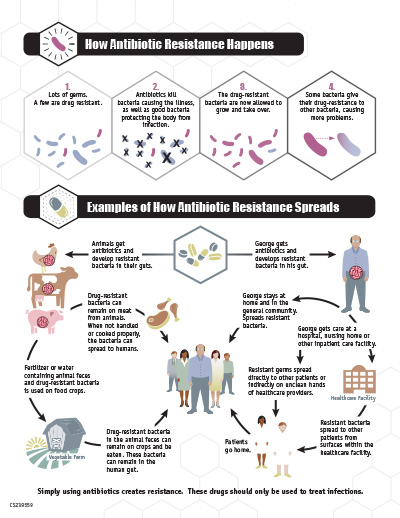
- Epidemiologic, traceback, and laboratory findings have linked this outbreak to contact with dairy bull calves purchased from livestock markets in Wisconsin. Dairy bull calves are young, male cattle that may be raised for meat.
- The Wisconsin State Laboratory of Hygiene and CDC’s National Antimicrobial Resistance Monitoring System (NARMS) laboratory conducted antibiotic-resistance testing on clinical isolates collected from ill people infected with an outbreak strain of Salmonella Heidelberg.
- All isolates were multidrug resistant.
- Antibiotic resistance may be associated with increased risk of hospitalization, development of a bloodstream infection, or treatment failure in patients.
- Follow these steps to prevent illness when working with any livestock, such as dairy bull calves:
- Always wash your hands thoroughly with soap and water right after touching livestock, equipment for animals, or anything in the area where animals live and roam. This is especially important to do before preparing or consuming food or drink for yourself or others.
- Use dedicated clothes, shoes, and work gloves when working with livestock. Keep and store these items outside of your home.
- Work with your veterinarian to keep your animals healthy and prevent diseases.
- This investigation is ongoing. This outbreak is a reminder to use a One Health approach to preventing illness, which recognizes that the health of people is connected to the health of animals and the environment.
Initial Announcement
November 28, 2016
CDC is working with Wisconsin health, agriculture, and laboratory agencies, several other states, and the U.S. Department of Agriculture Animal and Plant Health Inspection Service (USDA-APHIS) to investigate a multistate outbreak of multidrug-resistant Salmonella Heidelberg infections.
Public health investigators used the PulseNet system to identify illnesses that may have been part of this outbreak. PulseNet, coordinated by CDC, is the national subtyping network of public health and food regulatory agency laboratories. PulseNet performs DNA fingerprinting on Salmonella bacteria isolated from ill people by using techniques called pulsed-field gel electrophoresis (PFGE) and whole genome sequencing (WGS). CDC PulseNet manages a national database of these DNA fingerprints to identify possible outbreaks.
Twenty-one people infected with an outbreak strain of Salmonella Heidelberg have been reported from eight states. A list of states and the number of cases in each can be found on the Case Count Map page.
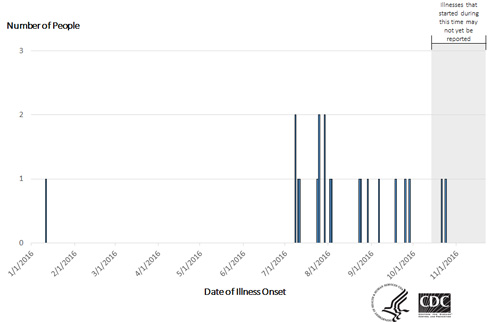
Among 19 people with available information, illnesses started on dates ranging from January 11, 2016 to October 24, 2016. Ill people range in age from less than 1 year to 72, with a median age of 21. Sixty-two percent of ill people are female. Among 19 ill people with available information, 8 (42%) reported being hospitalized, and no deaths have been reported.
WGS showed that isolates from ill people are closely related genetically to one another. This close genetic relationship means that people in this outbreak are more likely to share a common source of infection.
This outbreak can be illustrated with a chart showing the number of people who became ill each day. This chart is called an epidemic curve or epi curve. Illnesses that occurred after October 24, 2016 might not be reported yet because reporting takes an average of 2 to 4 weeks. Please see the Timeline for Reporting Cases of Salmonella Infection for more details.
Investigation of the Outbreak
Epidemiologic, traceback, and laboratory findings have identified dairy bull calves from livestock markets in Wisconsin as the likely source of infections. Dairy bull calves are young, male cattle that have not been castrated and may be raised for meat. Dairy bull calves in this outbreak have also been purchased for use with 4-H projects.
In interviews, ill people answered questions about any contact with animals and foods eaten in the week before becoming ill. Of the 19 people interviewed, 15 (79%) reported contact with dairy bull calves or other cattle. Some of the ill people interviewed reported that they became sick after their dairy bull calves became ill or died.
One ill person’s dairy calves were tested for the presence of Salmonella bacteria. This laboratory testing identified Salmonella Heidelberg in the calves. Further testing using WGS showed that isolates from ill people are closely related genetically to isolates from these calves. This close genetic relationship means that the human infections in this outbreak are likely linked to ill calves.
As part of routine surveillance, the Wisconsin State Laboratory of Hygiene, one of seven regional labs affiliated with CDC’s Antibiotic Resistance Laboratory Network, conducted antibiotic resistance testing on clinical isolates from the ill people associated with this outbreak. These isolates were found to be resistant to antibiotics and shared the same DNA fingerprints, showing the isolates were likely related to one another.
WGS identified multiple antimicrobial resistance genes in outbreak-associated isolates from fifteen ill people and eight cattle. This correlated with results from standard antibiotic resistance testing methods used by CDC’s National Antimicrobial Resistance Monitoring System (NARMS) laboratory on clinical isolates from two ill people in this outbreak. The two isolates tested were susceptible to gentamicin, azithromycin, and meropenem. Both were resistant to amoxicillin-clavulanic acid, ampicillin, cefoxitin, ceftriaxone, chloramphenicol, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim-sulfamethoxazole and had reduced susceptibility to ciprofloxacin. Antibiotic resistance limits treatment options and has been associated with increased risk of hospitalization, bloodstream infections, and treatment failures in patients.
Traceback information available at this time indicates that most calves in this outbreak originated in Wisconsin. Wisconsin health and agriculture officials continue to work with other states to identify herds that may be affected.
This investigation is ongoing. CDC will provide updates when more information is available.
More Information
- Page last reviewed: November 28, 2016
- Page last updated: November 28, 2016
- Content Source:


 ShareCompartir
ShareCompartir