Bioinformatics
GWAS Pipeline
Our genome-wide association study (GWAS) analysis package, Glu-Genetics, includes a set of command-line accessible modules that support many of the typical GWAS analysis requirements such as raw genotype conversion, processing, generation of summary statistics, analytical QC, fitting association models, aiding in interpretation of results, and follow-up study design.
Data Management
- Read, subset, transform, filter and merge tabular text and genotype files.
- Import low-level Illumina data for genotype and CNV analysis.
Genotype QC
- Compute completion rates by sample/locus, sample heterozygosity, and deviation from Hardy-Weinberg proportions.
- Test genotype concordance to verify duplicate samples and to detect unexpected duplicate samples.
- Verify reported sex.
- Compute IBS and IBD sharing to detect close relatives.
- Detect non-Mendelian inheritance patterns.
Population structure
- Estimate population admixture proportions and identify population outliers.
- Perform principal components analysis on large-scale genetic correlations to detect outliers and estimate adjustors for population stratification.
Association testing
- Linear regression models for quantitative traits.
- Logistic regression models including polytomous regression.
- Support for multiple genetic models, including unconstrained, trend, dominance, and recessive.
- Support for arbitrary categorical and continuous covariates and arbitrary interactions among covariates and genetic effects (GxG, GxE, ExE, etc.)
- Imputation-based analysis.
Genomic annotation
- Annotate SNPs with chromosome, cytoband, location, and nearby genes.
- Find SNPs near genes or regions.
LD estimation and tagging
- Generate pairwise LD values.
- Filter SNPs based on an LD threshold.
- Find LD surrogates for SNPs.
- Evaluate coverage of tagged SNPs.
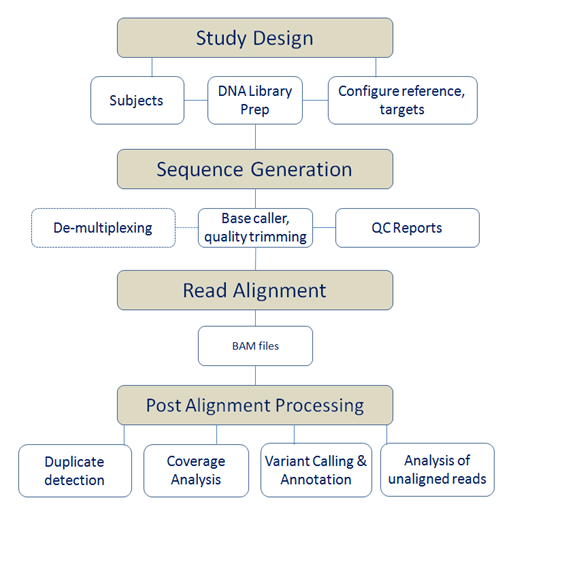
NGS Pipeline
Next-generation sequencing (NGS) platforms pose many challenges for data management, processing, and analysis. Our goals in addressing these challenges are two-fold: to standardize data management functions across multiple platforms and to make NGS data processing automatic, transparent, efficient, and reproducible. The current NGS pipeline supports multiple sequencing project designs, including short amplicon regional targets, long range amplicon regional targets, off-the-shelf and custom sequence-capture arrays from Nimblegen, Agilent, and RainDance, and a small amount of viral sequencing.