Biological Sciences Division
Research Highlights
January 2011
Sequenced Genomes Make Good Neighbors
Comparing mass spectra among organisms enables protein identification
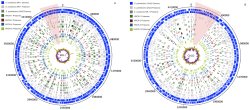
Protein identifications from Columbia River isolates are mapped to the reference genomes of S. oneidensis MR-1 (A) and S. putrefaciens CN32 (B). While all organisms were grown under the same conditions, observation of no protein expression compared to the reference proteome reveals these organisms have undergone evolutionary divergence. The protein identifications for each of the Shewanella species mapped onto their respective genomes, as well as the protein orthologs across species, also are shown. Two regions of "missing" proteome information from the Hanford Reach isolates are highlighted. Enlarge Image
Results: To study the proteomes of organisms, a first step often involves using sequenced genomes in conjunction with mass spectrometric measurements for global protein identifications. But, how do you identify the proteins in an organism yet to be sequenced? One way is to look at its sequenced neighbors, which is what scientists at Pacific Northwest National Laboratory (PNNL) did. They demonstrated a trans-organism search strategy for determining the extent to which near-neighbor genome sequences can be effective for global protein identifications in unsequenced organisms isolated from environmental samples.
In this strategy, mass spectra from an unsequenced organism were searched against the genome sequences for progressively more genetically distant neighbor organisms to determine how much proteome information could be obtained about one species when using the genomic sequence of another. The work appeared in PLoS ONE in November 2010.
Why it matters: The ability to identify the proteins in an organism provides scientists with information about an organism's physiological status, insights into gene function, and indicates which genes are activated and translated into functional proteins as organisms and microbial communities develop or respond to environmental cues.
Such information is important in developing a predictive understanding of biological systems relevant to energy production, environmental remediation, and climate change mitigation. However, despite the development of increasingly high-throughput sequencing technologies, sequenced genomes for every microorganism, particularly microbial communities, are not available.
Methods: The scientific team, led by PNNL scientists Dr. Mary Lipton and Dr. Stephen Callister, selected multiple genome sequences for Shewanellae to test the concept, not only because of the large number of publicly available genome sequences for this genus, but also the potential environmental importance of Shewanella organisms. They also included sequences from two other bacteria, Deinococcus radiodurans R1 and Salmonella typhimurium LT2, both distantly related (based upon 16s rDNA genes) to Shewanellae.
The team identified 300-500 proteins in four uncharacterized Shewanellae isolated from sediments sampled along the Hanford Reach of the Columbia River in Washington state and demonstrated an empirical relationship between the numbers of proteins identified. They did this using a neighboring sequenced genome to interpret mass spectrometric data and determine how closely related the sequenced organism was to the uncharacterized Shewanella organisms anticipated to be present in the sediments.
The range of identified proteins revealed that while a near-neighbor organism strategy can provide protein information for characterizing the proteomes of environmental isolates, selection of the near-neighbor organism is important because even organisms within the same genus can yield varying results. With careful selection of a closely related near neighbor, the application of this strategy potentially is useful for proteomics screening of environmental isolates.
What's next: Dr. Lipton and Dr. Callister are applying this strategy to study proteomes of microbial communities for which metagenomic information is not available.
Acknowledgments: The research was supported by the Department of Energy Office of Biological and Environmental Research (DOE-BER), the National Institute of Allergy and Infectious Diseases, and the National Institutes of Health's National Center for Research Resources. A portion of the research was performed using EMSL, a national scientific user facility sponsored by DOE-BER and located at PNNL.
Reference: Turse JE, MJ Marshall, JK Fredrickson, MS Lipton, and SJ Callister. 2010. "An empirical strategy for characterizing bacterial proteomes across species in the absence of genomic sequences." PLoS ONE 5(11):e13968. DOI: 10.1371/journal.pone.0013968.
