


Radio transripts from the 1960s reveal how a relatively new technology, the computer, had begun to alter research and the practice of medicine. “We have made extensive use [of computers] so far, but I think much more lies in the future,” said then NCI Director Dr. Carl G Baker during one such interview. “Massive amounts of information accumulate very rapidly.” Read more > >
The director of NCI's Center for Biomedical Informatics and Information Technology talks about caBIG®, which provides bioinformatics infrastructure and a portfolio of more than 40 tools that enable organizations and individual researchers to securely share biomedical data. Read more > >
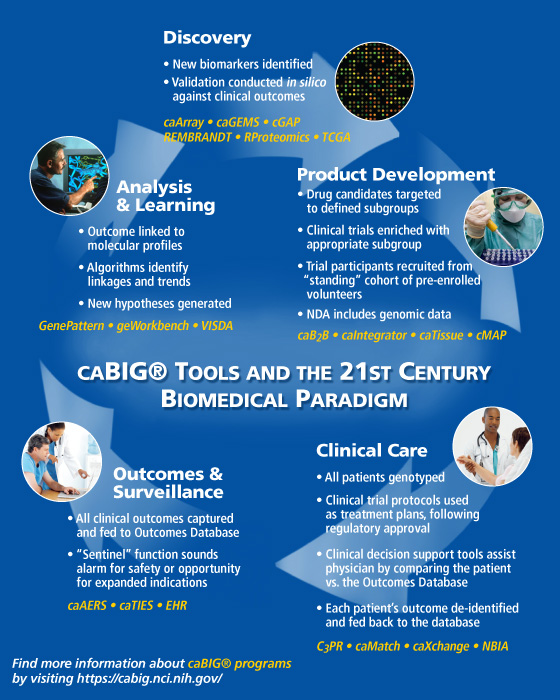
This diagram illustrates how the tools that have been developed through NCI's caBIG® project will enable the seamless integration of data from bench to bedside, making cancer research and patient treatment more efficient, and realizing the benefits of personalized medicine.

Top federal health information technology officials are predicting that the American Recovery and Reinvestment Act of 2009 will help push development and adoption of health IT and interconnectivity to dramatically new levels over the coming years. Read more > >
-
Computational algorithms are being used to organize and sift through the recent explosion of genomic information about tumors
-
Electronic health record systems are beginning to demonstrate their utility in research, and NCI is collaborating with ASCO to develop tools for more widespread adoption
-
In silico research can provide substantial time and cost savings to researchers by highlighting the most promising avenues for future research
-
Resources for funding, collaboration, and guidance can be found throughout NCI's divisions, as well as the Department of Health and Human Services

-
Common genetic changes may disable a protein that could block tumors
-
Profiling the genes HER2 and TOP2A could help guide the selection of therapies
-
Evidence further implicates JAK genes in acute lymphoblastic leukemia
-
Preliminary results show the drug cut risk by 23 percent compared to placebo
-
Growing populations of older adults and minorities will drive increase in the U.S.
-
Therapeutic vaccine shows modest benefit in phase III trial

-
- New HHS Secretary Sworn In
- NCI's Lowy Elected to NAS
- NCI's Ross Wins 2009 Kretchmer Award
- CCR Eminent Lecture Series Features Dr. Andrew Fire
- Telephone Workshop Series for Cancer Survivors
- DCLG Accepting Nominations
- NCI to Highlight Translational Research Resources at 2009 BIO International Convention
The NCI Cancer Bulletin is produced by the National Cancer Institute (NCI), which was established in 1937. Through basic, clinical, and population-based biomedical research and training, NCI conducts and supports research that will lead to a future in which we can identify the environmental and genetic causes of cancer, prevent cancer before it starts, identify cancers that do develop at the earliest stage, eliminate cancers through innovative treatment interventions, and biologically control those cancers that we cannot eliminate so they become manageable, chronic diseases.
For more information about cancer, call 1-800-4-CANCER or visit http://www.cancer.gov.
NCI Cancer Bulletin staff can be reached at ncicancerbulletin@mail.nih.gov.
 |

Director's Update: Enabling the Evolution of Oncology
 Dr. John E. Niederhuber
Dr. John E. Niederhuber
Radio transcripts from the 1960s reveal how a relatively new technology, the computer, had begun to alter research and the practice of medicine. "We have made extensive use [of computers] so far, but I think much more lies in the future," said then NCI Director Dr. Carl G. Baker during one such interview. "Massive amounts of information accumulate very rapidly."
Dr. Baker's comments hinted at what we see today, a time when journal publications and research conferences regularly reveal new ways to identify the nuances of a person's biological profile that may help tailor interventions for cancer to maximize clinical benefit and mitigate potential harm. We are entering the age of personalized medicine.
However, a robust bioinformatics infrastructure is critical for achieving truly individualized care. That means developing the tools necessary to mine and analyze the vast amounts of clinical, molecular, and epidemiologic data.
I'm proud to say that we are moving in that direction. For example, using an approach whereby glioblastoma (GBM) tumor samples were painstakingly characterized using different genomic strategies, researchers with The Cancer Genome Atlas (TCGA) identified new mutations, a core set of often simultaneously deregulated molecular pathways, and a potential mechanism of resistance to the most commonly used chemotherapy agent for GBM. In effect, three actionable findings from a sophisticated, collaborative analysis that could eventually lead to more tailored treatments for patients with this disease.
TCGA, like many other advances reported on regularly in the NCI Cancer Bulletin, would not exist without the underlying technology that enables these discoveries. That is why we are highlighting in this special issue of the NCI Cancer Bulletin the role of bioinformatics in cancer research and issues that surround its development and adoption.
 Computers from the 1960s
Computers from the 1960s
At NCI, we are investing heavily in developing a nationwide bioinformatics structure, most notably through our Center for Biomedical Informatics and Information Technology and our landmark initiative, the cancer Biomedical Informatics Grid, or caBIG®. Our goal is to create a seamless system for the storage and access of data—a system that facilitates data exchange and collaboration, so that a cancer center in California can easily interface with a community oncology center in Tennessee. In this way, the same data a community oncologist uses to work with a patient in making a treatment decision can also be used by researchers analyzing whether a new screening test improves survival.
A key element in this bioinformatics equation will eventually be electronic health records (EHRs). President Obama and his administration have directed a significant portion of ARRA funding for health information technology, including EHRs, and NCI staff in various divisions are working on important EHR-related projects.
The topic of bioinformatics is remarkably broad. I encourage you to follow up with our staff and the programs listed in the following articles to find out more about their tools and projects and how you can get involved.
Dr. John E. Niederhuber
Director, National Cancer Institute
 |

A Conversation with Dr. Kenneth Buetow
 Dr. Kenneth Buetow
Dr. Kenneth Buetow
Director of NCI's Center for Biomedical Informatics and Information Technology and leader of the caBIG® (cancer Biomedical Informatics Grid) initiative, which provides bioinformatics infrastructure and a portfolio of more than 40 tools that enable organizations and individual researchers to securely share biomedical data.
How has caBIG® addressed interoperablity with the proprietary systems in place at some institutions?
There are two paths to connect with caBIG®: "adoption" and "adaptation." Many institutions choose a hybrid approach of adopting some caBIG® tools and adapting some of their existing IT infrastructure using published compatibility guidelines to become caBIG®-compliant. All caBIG® tools and information are released under a "non-viral" open-source license that allows commercial reuse of caBIG® technology, so that vendors may become caBIG®-compatible. We have adopted internationally recognized data standards or worked with professional organizations to develop vocabularies, common data elements, and data models where such standards did not exist or were inadequate, simplifying our ability to connect with other systems. Because caBIG® supports data federation, organizations maintain control over their own data. We recently demonstrated health data exchange between systems at the Department of Defense and Kaiser Permanente using interoperable caBIG® clinical trials management software, bridging what has traditionally been perceived as an insurmountable chasm between research and care.
How are issues surrounding information security addressed?
The Data Sharing and Intellectual Capital Workspace of caBIG® addresses this issue, and provides guidelines and tools to help researchers evaluate the sensitivity of their data and address federal privacy regulations, human participant protections, sponsor contract compliance, and proprietary interests. In addition, the caGrid architecture is based on the open-source Globus toolkit with additional enhanced security infrastructure that provides services and tools for the administration and enforcement of security policy.
What is next for caBIG®?
The past year has been one of huge accomplishments for the caBIG® program. Fifty NCI-designated Cancer Centers and members of the NCCCP have connected to each other via caGrid, creating the world's largest dedicated biomedical research grid, with more than 120 active grid nodes. To support the continued growth of the community, the Enterprise Support Network was established, composed of six Knowledge Centers, each supporting a scientific domain or collection of caBIG® tools. In addition, Support Service Providers—almost 20 to date—assist researchers and organizations who are connecting to caBIG® and require training, installation, or tool customization, on a fee-for-service basis.
Beyond the United States, caBIG® and the National Cancer Research Institute in the UK have been working together and adopting complementary technologies for years. Several caBIG® members recently traveled to India to meet with representatives of the health ministry, leading research hospitals, and the Center for the Development of Advanced Computing. We look forward to many more productive interactions in the future. In a vote of confidence for the quality and usefulness of caBIG® technology, the King Hussein Medical Center in Amman, Jordan, recently decided to implement caBIG® infrastructure across the entire hospital to provide data interoperability.
For more information on the caBIG® program, I encourage everyone to read the recently published caBIG® 2008 Annual Report at http://cabig.cancer.gov/gettingconnected/caBIGresources/annualreport/.
 |

Recovery Act Boosts Bioinformatics
Top federal health information technology (IT) officials are predicting that the American Recovery and Reinvestment Act of 2009 (ARRA) will help push development and adoption of health IT and interconnectivity to dramatically new levels over the coming years. NCI's pioneering cancer Biomedical Informatics Grid® (caBIG®) project—and its current and prospective partners in the cancer research community—are likely to benefit substantially from the infusion of federal funding and new policies fostering these national economic recovery goals.
"With ARRA we finally have significant resources to build on the foundation that's been created for health IT and take this to the next level," said Kelly Cronin, director of the Office of Programs and Coordination at the HHS Office of the National Coordinator for Health Information Technology (HIT), during the April 16 World Health Care Congress in Washington, DC.
She noted that ARRA provides two "buckets" of federal funding for developing health bioinformatics. The first bucket contains $2 billion in discretionary funds for HHS programs that foster health care workforce development and state grants for building health IT infrastructure. HHS will also fund regional extension centers and a national research center to provide technical assistance and implementation support for widespread adoption of health IT systems.
The second bucket provides about $18 billion in incentive payments over 10 years, starting in 2011, through Medicare and Medicaid for physician practices and hospitals to become "meaningful users" of health IT and patient electronic health records. This includes incentives for providers to join the proposed Nationwide Health Information Network (NHIN) and share information across the network.
Dr. Kenneth Buetow, director of NCI's Center for Biomedical Informatics and Information Technology, said, "NCI and caBIG® have been very active participants with the federal efforts to create the NHIN. When the first prototype of NHIN was demonstrated last December, we showed that caBIG® could interconnect with the national network. We're excited and optimistic about NCI's role and the role of the cancer community overall in this opportunity to be on the leading edge of the deployment of electronic health systems across the country."
Among NCI's strategic focus for caBIG® in the next few years, Dr. Buetow explained, two activities stand out. First, NCI will guide the full-scale deployment of caBIG® among the NCI-designated Cancer Centers. NCI has worked closely with the Centers over the past 5 years to create the technological infrastructure and interconnected suites of research tools that make up caBIG®.
"The second strategic activity that's new to caBIG® is bringing the newly minted NCI Community Cancer Centers Program (NCCCP) into the network," Dr. Buetow continued. The 16 sites participating in the NCCCP pilot program have agreed to interconnect through caBIG®. "We believe that the investment by NCI is beginning to pay off with large-scale deployments of caBIG®," he added. "Now with the ability to connect much more directly through NCCCP to the sites of primary health care delivery, we'll be in a position to explore a much more efficient means of conducting clinical research and interconnecting with the community for primary oncology care delivery."
 |

Cancer Genomics: Building Haystacks, Finding Needles
Without computers and sophisticated mathematics to organize and sift through the recent explosion of genomic information about tumors, important clues to cancer might have remained hidden within jumbles of genetic code.
But of course this has not happened. Instead, with increasing efficiency over the last decade, biological information has been gathered, stored on computer servers, and shared through the Internet. With the help of informatics, researchers around the world have been mining this repository of data and uncovering more than a few new cancer-related findings.
 Claude Monet “Grainstack (Sun in the Mist)” 1891, Oil on canvas Minneapolis Institute of Arts
Claude Monet “Grainstack (Sun in the Mist)” 1891, Oil on canvas Minneapolis Institute of Arts
One surprise was the recent discovery of fused genes in prostate cancer. Gene fusions, which arise when DNA sequences from two genes merge inappropriately, are a hallmark of cancers of the blood. But they had eluded detection in "solid" tumors until 2005, when a bioinformatics approach was applied to the challenge.
Researchers at the University of Michigan Medical School developed an algorithm to search an online database called Oncomine for unusual patterns of gene activity in subsets of prostate tumors. It's now known that fused genes are common in prostate tumors and may drive the disease. Fused genes have also been found in lung tumors and may yet be discovered in other common cancers.
Like many online databases, Oncomine can be used to explore diverse questions about cancer biology, and it could not have been built in the days before bioinformatics. The database contains results from 20,000 microarray experiments, most of which collected information on thousands of genes.
Another such bioinformatics resource is the Connectivity Map. Developed at the Broad Institute and supported by NCI's Integrative Cancer Biology Program, this is an online database of gene signatures that can help identify potential drugs for treating disease. Users can search for drugs that modify the genetic program of a cancer cell in a way that may benefit patients. Recent studies have yielded candidates for targeting leukemia stem cells and treating a rare leukemia.
As with all such computational predictions, the findings need validation. Nonetheless, bioinformatics can provide leads when there are few other options. For example, computational algorithms have uncovered what are truly needles in the haystack of the human genome—microRNAs. First identified in other species, these snippets of genetic material are only about 22 nucleotides in length. But they are important regulators of genes and have been linked to cancer and metastasis.
Bioinformatics can also help with a central challenge of cancer genomics: Distinguishing genetic alterations that initiate and fuel cancers (known as drivers) from changes that are merely present in tumors but do not contribute to the disease (the passengers). Computational tools can help identify potential drivers based on statistical measures such as how common a mutation is.
With all studies of cancer genomes, the goal is always to translate knowledge into improvements in the prevention, detection, and treatment of the disease. Being able to collect and compare large amounts of clinical and genomic data can help achieve this goal, as a recent finding from The Cancer Genome Atlas (TCGA) project suggests.
By comparing genomic and epigenomic data on brain tumors with the treatment records of patients, the study uncovered a potential mechanism of resistance to a cancer drug. While the insight itself is perhaps not unusual, the promise of bioinformatics is that by making such comparisons on a large scale and with powerful analytical tools, scientists can accelerate the pace of discovery.
 |

Electronic Health Records Emerging as Important Care,
Research Tool
Bringing EHRs to the Community
Because of their potential to improve care and clinical research, NCI and the American Society of Clinical Oncology (ASCO) are collaborating to bring EHRs to community hospitals and centers where most cancer patients receive their care. Under the collaboration, NCI and ASCO are establishing specifications for an oncology-specific EHR that will rely on caBIG® standards for interoperability.
The specifications, expected to be completed later this year, are being developed based on open standards in use in the oncology community. The EHRs developed based on these specifications, which may come from industry, NCI, or both, will then be deployed to participants in NCI's National Community Cancer Center Program.
With the American Recovery and Reinvestment Act set to spur their development and implementation, electronic health records (EHRs) are getting a lot of attention while businesses like Google and Wal-Mart have begun to develop their own EHR tools.
The widespread adoption of EHRs, however, involves "huge challenges," acknowledged Dr. David Blumenthal, the National Coordinator for Health Information Technology. As a recent study he led documented, less than 2 percent of U.S. hospitals have a comprehensive EHR system in place. Cost, the study found, was the biggest obstacle to adoption.
Despite some of the problems reported to date with EHRs, evidence is emerging that they can improve the quality and efficiency of medical care. For example, the relatively new EHR system at the University of Arkansas for Medical Sciences (UAMS) has made many aspects of delivering care “so much better,” said Dr. Laura Hutchins, director of Hematology/Oncology at the UAMS Winthrop P. Rockefeller Cancer Institute. While the system is not perfect, she continued, "I don't know of anybody here who wants to go back to a paper record. In addition to saving money, she explained, the system has generally made patient visits more efficient—for example, streamlining the search for information that can influence diagnosis or treatment.
Whereas the UAMS system is still in its early days, the EHR system at the University of Pittsburgh Medical Center (UPMC) dates back to 1991. The center recently completed the first phase of an "interoperability initiative" intended to eventually provide staff at 20 hospitals and more than 400 physician offices and outpatient sites access to what Dr. Daniel Martich, UPMC's chief medical information officer, calls a "full-fidelity" EHR system, an integrated network of patient records with data on everything from admissions to allergies to recent imaging studies. While access to a number of EHR-related tools, such as electronic prescribing, still varies, he explained, the goal is a widely accessible EHR system that "provides a unified view of what's going on with the patient."
Importantly, EHR systems are beginning to demonstrate their utility in research. At UPMC, for example, they have conducted studies showing that, with the addition of clinical prompts, the EHR system reduced the risk of patients receiving an overdose of acetaminophen and improved by fivefold the number of patients notified by their primary care physicians that they may be candidates for clinical trials. Dr. Hutchins and colleagues at UAMS, meanwhile, used their EHR system to evaluate vitamin D levels in women with metastatic breast cancer who received bisphosphonates to treat bone pain and osteoporosis, finding that vitamin D supplements were being underprescribed, which can affect patient outcomes.
The success of EHRs, Dr. Martich believes, will be measured by the extent to which they can be effectively integrated into clinical care and research systems. "The real issue [with EHRs] isn't a technological one," he said. "The question is: How do they function within the workflow of a health care system?"
 |

Computational Modeling Paints a Picture of the Future
 Researchers at the Harvard-MIT Complex Biosystems Modeling Laboratory (Massachusetts General Hospital), which is supported by NCI's Integrative Cancer Biology Program, have developed a virtual model that predicts the growth of brain tumors over time (shown here at time step 52 and 110), taking into consideration EGFR gene-protein interactions and the effect of glucose and oxygen concentrations on cell movement and division. Models such as this one can be used to integrate data and generate hypotheses for tumorigenesis, cancer detection and treatment. (Image courtesy of Dr. Thomas Deisboeck, learn more at http://biosystems.mit.edu and https://www.cvit.org)
Researchers at the Harvard-MIT Complex Biosystems Modeling Laboratory (Massachusetts General Hospital), which is supported by NCI's Integrative Cancer Biology Program, have developed a virtual model that predicts the growth of brain tumors over time (shown here at time step 52 and 110), taking into consideration EGFR gene-protein interactions and the effect of glucose and oxygen concentrations on cell movement and division. Models such as this one can be used to integrate data and generate hypotheses for tumorigenesis, cancer detection and treatment. (Image courtesy of Dr. Thomas Deisboeck, learn more at http://biosystems.mit.edu and https://www.cvit.org)
In silico research (better known as computational modeling or mathematical modeling) uses complex algorithms requiring high-powered computers to predict unknown properties or outcomes of disease.
Modeling is a familiar tool in the cancer research arsenal, explained Dr. Daniel Gallahan, director of NCI's Integrative Cancer Biology Program (ICBP): "After all, the mouse is a model system. It's a very complex system—we don't know all the components that go into making a mouse—but it's a sophisticated model in that it mimics a lot of the cellular and molecular processes that are ongoing in human cancer."
The ICBP's computational models aim to mimic the most fundamental elements of cancer, "for example, looking at chemical interactions, or molecules that interact within a cell and cause malignant transformation," said Dr. Gallahan. "It can be very efficient to run a computational program as opposed to setting up a series of biological experiments."
Any model's predictions must be validated in a biological system, but modeling can provide substantial time and cost savings to researchers by highlighting the most promising avenues for future research. Nine centers funded by the ICBP are currently building predictive models of a wide range of biological processes related to carcinogenesis and cancer treatment, including cell signaling pathways, epigenetic changes, and response to targeted therapies.
On the opposite end of the cancer research continuum, NCI's Cancer Intervention and Surveillance Modeling Network (CISNET) uses computational modeling to understand how cancer control interventions influence the disease at the population level. "We observe trends in the national cancer rate, and then try to understand why they're occurring," explained Dr. Eric Feuer, program director of CISNET.
The complex factors that influence national cancer trends are virtually impossible to study in a controlled fashion in the real world. "What modeling does is let us build a sort of virtual world that allows us to decompose the real world into the components that influence these trends," said Dr. Feuer.
For example, after many randomized controlled trials and meta-analyses of mammography, there was still controversy about whether or not mammography reduces mortality from breast cancer. In 2005, using seven independent mathematical models, CISNET investigators were able to show that declines in U.S. breast cancer mortality observed from 1975 to 2000 would be very difficult to explain without a substantial contribution from mammography.
The ICBP and CISNET have recently begun pilot collaborations to model several cancer types all the way from their cellular biology to population-level effects, with the ultimate goal of being able to accurately simulate the results of clinical treatment trials.
“I think the 'holy grail' of modeling, or even understanding cancer, is to have a unified theory—to be able to measure what's going on within a cell at the molecular level, then predict all the way through to what would occur in a population," explained Dr. Gallahan. "It's daunting, but if you look at the specific aspects of it, we're not trying to conquer the world in one fell swoop; we're simply trying to tease it apart, and that's the power and possibility of modeling.”
 |

Learn More
| NCI Collaborations, Tools, and Funding Opportunities |
| Other Resources and Funding Opportunities |
 |

Mutant Protein Implicated in Diffuse Large B-cell Lymphoma
Recent studies have implicated the NF-κB signaling pathway in the development of a subset of diffuse large B-cell lymphomas. Two new studies now provide another piece of the puzzle. While mutations in genes such as CARD11 can spur cell growth by activating the NF-κB pathway, mutations in a gene called A20 may remove a natural "brake" on the pathway. The findings suggest that multiple lesions in the NF-κB pathway may be involved in DLBCL, the most common lymphoma in adults, according to results published online in Nature this week.
"We have identified genetic lesions that target multiple components of the same pathway in the majority of DLBCL with constitutive activation of NF-κB, and this will have implications for the possible design of therapies that may benefit patients with these abnormalities," said Dr. Laura Pasqualucci of the Herbert Irving Comprehensive Cancer Center at Columbia University, who led one of the studies. Drugs targeting the NF-κB pathway are in development, she noted.
Her team analyzed tumor samples from 168 cases of DLBCL. Mutations in multiple genes associated with the NF-κB pathway were found in half of the activated B-cell-like (ABC) subtypes analyzed, and in a smaller fraction of the germinal center B-cell-like (GBC) subtypes. The A20 gene was the most commonly altered, with a third of the ABC-DLBCL patients showing inactivation of both gene copies by mutation or deletion.
In the second study, researchers at the University of Tokyo conducted genome-wide analyses of genetic lesions in 238 B-cell lymphomas. The A20 protein was frequently inactivated in several types of cancer, including mucosa-associated tissue lymphoma, a form of Hodgkin's lymphoma, and to a lesser extent, B-cell lymphomas. Both teams found that, in laboratory experiments, the normal A20 protein suppressed cell growth and caused abnormal cells to commit suicide when reintroduced into A20-deficient cells.
Testing Breast Tumors May Predict Response to Chemotherapy
Women treated for breast cancer whose tumors carry normal versions of the genes HER2 and TOP2A may not benefit from an anthracycline as part of additional chemotherapy designed to prevent a recurrence. Instead, these patients may benefit from a less toxic regimen that does not include an anthracycline, researchers reported online in the Journal of the National Cancer Institute on April 28.
In previous studies, the benefits of adjuvant therapy with an anthracycline-based regimen were restricted to women with HER2 alterations (about 20 percent of breast cancers). But because the TOP2A gene resides near HER2, some researchers have wondered whether the response to anthracyclines might be associated with TOP2A alterations.
To explore the question, Dr. Kathleen Pritchard of Sunnybrook Odette Cancer Centre in Toronto and her colleagues analyzed these genes in tumor samples from 438 of the 710 participants in the National Cancer Institute of Canada's Mammary 5 trial.
Women whose tumors had either TOP2A deletions or amplifications (extra copies) had longer recurrence-free survival and overall survival in response to chemotherapy that included the anthracycline epirubicin than to chemotherapy without anthracyclines, while patients with normal TOP2A genes showed no difference in responsiveness. Alterations in TOP2A and in HER2 appear to have similar value in guiding the selection of anthracycline-containing regimens, the researchers concluded, noting that larger studies are needed to determine which measurement is more closely associated with response to these regimens.
An accompanying editorial agrees that women whose tumors have normal HER2 and TOP2A genes should not receive anthracycline-based chemotherapy. The authors note that molecularly targeted drugs such as trastuzumab (Herceptin) often provide the most benefit to patients with alterations in the pathways affected by the drugs, and it now appears that the same may be true of standard chemotherapy agents.
More Gene Mutations Found in Childhood Leukemia
In January, researchers described a new subtype of acute lymphoblastic leukemia (ALL), the most common childhood cancer. Children with this subtype have alterations to a gene called IKAROS (or IZKF1) and a high risk of relapse. The researchers predicted that some cases would also involve mutations in protein kinase genes, which play a role in cell signaling and are commonly altered in cancer. New research now confirms that prediction.
A genetic analysis of 187 cases of high-risk childhood ALL by investigators with the Childhood Cancer TARGET Initiative has identified mutations in three members of the family of JAK kinase genes. The JAK mutations are thought to activate pathways involved in cell growth and proliferation. Such changes have also been implicated in other cancers, most notably in a group of blood cancers known as myeloproliferative disorders.
About 10 percent of the ALL cases had mutations in one of the three JAK genes. Some also involved changes to IKAROS, and these children had poor outcomes. More than 70 percent of children with both IKAROS and JAK mutations relapsed within 4 years, compared with 23 percent with neither alteration.
Drugs that inhibit overactive JAK kinase proteins are in development. Based on laboratory experiments, the researchers are hopeful that these agents might benefit patients with these mutations. In the lab, JAK mutations caused normal cells to become cancerous, while an experimental JAK inhibitor caused cells with the mutations to die.
The research team includes investigators from St. Jude Children's Research Hospital, the University of New Mexico Cancer Center, the Children's Oncology Group, and NCI. Dr. Charles Mullighan of St. Jude presented the findings on behalf of the TARGET team at the American Association for Cancer Research annual meeting last month in Denver.
Dr. Mullighan pointed out that JAK genes were mutated in children with ALL who did not also have Down syndrome. Several recent reports described mutations in the JAK2 gene among children who had both ALL and Down syndrome.
"The discovery of activating JAK mutations in a subset of ALL patients is a very important observation with obvious clinical implications," said Dr. Malcolm Smith of NCI's Cancer Therapy Evaluation Program and an NCI leader of the TARGET Initiative. The project is systematically sequencing more than 120 genes suspected of playing a role in ALL.
The incorporation of JAK inhibitors into treatment programs for patients with JAK-mutated ALL is a highly promising line of clinical research that needs to be aggressively pursued, added senior author Dr. Cheryl Willman of the University of New Mexico. She cited the success of imatinib (Gleevec), another kinase inhibitor, as a model.
The researchers are testing JAK inhibitors in their experimental models and hope eventually to move to patients. Genetic tests could be developed to screen patients for changes to the IKAROS and JAK genes, and the results could guide treatment as well as identify patients at risk of relapse, noted Dr. Mullighan.
Dutasteride May Reduce Prostate Cancer Risk
Initial data from a large, international clinical trial indicate that dutasteride (Avodart) may help prevent prostate cancer among men at higher risk for the disease, according to an April 27 report at the American Urological Association annual meeting in Chicago.
The trial, called REDUCE, compared dutasteride treatment against placebo among 8,200 men considered to be at high risk for the disease because of their elevated levels of prostate-specific antigen (PSA). All the men had received negative (clean) prostate biopsies within 6 months before joining the study.
After follow-up biopsies at 2 and 4 years, dutasteride was shown to lower the risk of prostate cancer by 23 percent compared with men taking the placebo.
Men treated with dutasteride were also found to be at no greater risk than those on placebo for developing aggressive prostate tumors. "We are very encouraged by this finding," lead investigator Dr. Gerald Andriole of the Washington University School of Medicine said in a statement. The study was funded by GlaxoSmithKline, which manufactures dutasteride.
The results are comparable to those from the NCI-funded Prostate Cancer Prevention Trial (PCPT), an earlier prevention study for prostate cancer involving a drug of the same class, finasteride (Proscar). Initial findings from PCPT suggested that finasteride decreased the risk of prostate cancer but may have increased the risk of developing more aggressive tumors. Subsequent investigations by NCI scientists and others showed that finasteride did not promote more aggressive tumors and may actually reduce their risk. Both finasteride and dutasteride are approved to treat benign prostatic hyperplasia.
Cancer Incidence Could Rise Sharply in Coming Decades
The number of cancer cases in the United States is expected to increase dramatically over the next 2 decades, particularly among older adults and minorities, according to a study published online last week in the Journal of Clinical Oncology (JCO). Dr. Ben Smith of the University of Texas M.D. Anderson Cancer Center and his colleagues used information from the U.S. Census Bureau and NCI's SEER database, which covers approximately 26 percent of the U.S. population, to project the number of cancer patients diagnosed through 2030 by various measures.
The total cancer incidence is projected to rise by about 45 percent, from 1.6 million in 2010 to 2.3 million in 2030, the study found. This will be driven largely by cancer diagnoses in growing populations of older Americans and minority groups. The study projects a 67 percent increase in cancer incidence among older adults, compared with an 11 percent increase for younger adults. A 99 percent increase is expected among minorities, compared with a 31 percent increase for whites.
Certain difficult-to-treat cancers, such as liver, stomach, pancreas, and lung, will likely be among those with the highest relative increases in incidence. Therefore, the study warns, unless substantial gains are made in the treatment and prevention of these diseases, particularly among the elderly and minorities, the number of cancer deaths could grow dramatically in the next 20 years.
A second article published online in JCO proposes a roadmap for addressing and overcoming disparities in cancer care. The authors of this policy statement, developed by the American Society of Clinical Oncology (ASCO), wrote that despite decades of investment and advances in cancer research, a "profound divide" exists between those with access to the fruits of this research and those without.
The paper outlines strategies for addressing health disparities, such as funding research on the quality of care provided to minority populations and boosting minority enrollment in clinical trials. The statement "sets the stage for the continuing activities by ASCO to address this very important problem," said ASCO president Dr. Richard L. Schilsky of the University of Chicago at a press briefing.
More information and audio files from the briefing are available on the ASCO Web site.
Immunotherapy Improves Survival in Metastatic Prostate Cancer
An investigational immunotherapy treatment improved overall survival by approximately 4 months in men with metastatic prostate cancer compared with men treated with a placebo, researchers reported last week at the American Urological Association annual meeting in Chicago. The results come from the IMPACT trial, a phase III, double-blind, randomized trial of sipuleucel-T (Provenge), a form of immunotherapy in which antigen-presenting cells are isolated from patients' blood, engineered to stimulate a tumor-specific immune response, and infused back into patients.
The more than 500 men in the trial had asymptomatic or minimally symptomatic, androgen-independent metastatic prostate cancer. In men who received sipuleucel-T, which was delivered in three infusions over a 1-month period, median survival was improved by 22.5 percent compared with men who received placebo (25.8 months versus 21.7 months). As was the case in the two earlier-stage trials of sipuleucel-T, there was no statistically significant improvement in progression-free survival, that is, survival without tumor growth.
Adverse events were minor and limited, said one of the trial's leaders, Dr. David Penson from the University of Southern California. The most common events were fever, chills, and headache the day after the infusion of sipuleucel-T, and these side effects typically resolved within a day or two. Overall, approximately 99 percent of patients in the immunotherapy arm received all three infusions.
The survival data were "incredibly consistent" in all of the subgroups examined in the trial, Dr. Penson explained, which included breakdowns by age, baseline PSA level, and extent of bone metastases, among others. "That is very reassuring to me," he said.
Dr. Penson acknowledged that, because of the trial's design, all patients in the immunotherapy arm could receive docetaxel immediately upon progression, unlike patients who received the placebo, and this could introduce bias in favor of sipuleucel-T. But the statistical model, he noted, was adjusted for both the use and timing of docetaxel administration. Further details of the data analysis should be available when the trial results are published in a scientific journal.
In March 2007, an FDA advisory committee recommended that sipuleucel-T be approved for men with this prostate cancer indication, based on data from two smaller clinical trials. In May 2007, however, the FDA issued a "complete response" letter to Dendreon, which manufactures sipuleucel-T, requesting more efficacy data before it could approve the company's application to market the treatment. According to Dendreon officials, the company will submit the IMPACT data to the FDA later this year as an amendment to its earlier marketing approval application.
 |

New HHS Secretary Sworn In
 Secretary Kathleen Sebelius
Secretary Kathleen Sebelius
Last week, Kathleen Sebelius was confirmed by the Senate and sworn in as the new Secretary of the Department of Health and Human Services (HHS) by President Barack Obama. Secretary Sebelius brings more than 20 years of experience in state government to her new role as head of one of the Federal government's largest departments, which includes NCI. More information about the new Secretary is available here.
NCI's Lowy Elected to NAS
 Dr. Douglas Lowy
Dr. Douglas Lowy
Last week, the National Academy of Sciences (NAS) elected to its ranks Dr. Douglas Lowy, chief of the Laboratory of Cellular Oncology in the Center for Cancer Research. The research of Dr. Lowy and his colleague Dr. John Schiller contributed to the commercial development of an HPV vaccine to prevent cervical cancer.
NAS members are selected in recognition of their distinguished and continuing achievements in original research. Election to the Academy is considered one of the highest honors that can be accorded to a U.S. scientist or engineer.
NCI's Ross Wins 2009 Kretchmer Award
 Dr. Sharon Ross
Dr. Sharon Ross
Dr. Sharon Ross of NCI's Division of Cancer Prevention (DCP) was recently awarded the prestigious Norman Kretchmer Memorial Award in Nutrition and Development from the American Society for Nutrition (ASN). The presentation ceremony took place during the ASN annual meeting in New Orleans on April 19.
Dr. Ross is currently a health scientist administrator and program director in DCP's Nutritional Science Research Group. Her research focuses on the epigenetic effects of nutrient availability on gene expression and cell development, both in embryology and carcinogenesis.
CCR Eminent Lecture Series Features Dr. Andrew Fire
 Dr. Andrew Fire
Dr. Andrew Fire
NCI's Center for Cancer Research (CCR) continues its Eminent Lecture Series presentations by nationally recognized scientists doing cutting-edge research.
Dr. Andrew Fire, Professor of Pathology and Genetics at Stanford University, will present the series' next lecture on May 18 at 3:00 p.m. in Lipsett Amphitheater on the NIH campus in Bethesda. The title of Dr. Fire's talk is "Tracking B Cell Diversity and Clonality in Human Immunity and Disease." Dr. Fire was jointly awarded the Nobel Prize in Physiology or Medicine in 2006, along with Dr. Craig Mello, for their discovery of RNA interference, gene silencing by double-stranded RNA.
The lecture series is free and open to the public. To learn more details about this and other upcoming lectures in the series, click here.
Telephone Workshop Series for Cancer Survivors
The seventh annual telephone workshop series "Living With, Through, and Beyond Cancer" continues on May 19 with part II of the series, entitled "The Importance of Nutrition and Physical Activity." This three-part series offers cancer survivors, their families, friends, and health care professionals practical information to help them cope with concerns and issues that arise after treatment ends.
The program is a collaborative effort between NCI, CancerCare, the Lance Armstrong Foundation, the Intercultural Cancer Council, Living Beyond Breast Cancer, and the National Coalition for Cancer Survivorship.
The workshops are free; no telephone charges apply. To register, visit the CancerCare Web site. The remaining workshops will take place from 1:30 p.m. to 2:30 p.m. EDT on the following dates:
Part II: "The Importance of Nutrition and Physical Activity," May 19
Part III: "Survivors Too: Family, Friends, and Loved Ones: Managing the Fatigue of Caregiving," June 23
The workshops are also archived (including Part I: "Managing the Stress of Survivorship") and available online as podcasts.
DCLG Accepting Nominations
NCI is currently identifying individuals to serve on the Director's Consumer Liaison Group (DCLG). The DCLG is a Federal Advisory Committee of 16 individuals who advise the NCI Director from the viewpoint of the consumer advocate. The Institute will be filling three positions on the board with dates of service beginning July 2009 and running through June 2013. All nomination materials must be received by NCI no later than 6:00 p.m. on May 22.
Further information about the DCLG and complete details about the nomination process can be found online. Previous nominees should be aware that in order to be considered for current vacancies, their updated materials must be resubmitted.
Please direct questions to ncidclgnominations@mail.nih.gov.
NCI to Highlight Translational Research Resources at 2009 BIO International Convention
While at the 2009 BIO International Convention, please visit the NCI exhibit at booth #3805 to learn about the many resources, partnerships, and collaborative opportunities available from NCI that enable the translation of cancer research discoveries to new cancer therapies and diagnostics. The NCI exhibit will feature a new interactive experience that provides an up-close look at how NCI's programs and initiatives are speeding the development of new diagnostic tests, cancer treatments and devices, and other interventions that benefit people with cancer and those who are at risk.
NCI experts will also be joining industry leaders in discussing critical topics in the life sciences during the following breakout sessions.
Tuesday, May 19
10:00 - 11:30 a.m. |
|
Transforming the Research Paradigm: 21st Century Models to Unify Discovery Research and Clinical Care
Ken Buetow, associate director for Biomedical Informatics and Information Technology, NCI Center for Biomedical Informatics and Information Technology |
| |
Wednesday, May 20
8:00 - 9:30 a.m. |
|
Fast Forwarding Life Science Innovation: What Works, What Doesn't, Where Do We Go From Here?
Carolyn Compton, director, NCI Office of Biorepositories and Biospecimen Research |
| |
Thursday, May 21
8:00 - 9:30 a.m. |
|
Private Companies Tackling Public Health
Michael Weingarten, director, NCI Small Business Innovation Research Development Center |
 |
|


















