 |
Fiscal Year 1999 President's Budget Request
Statement by
Dr. Francis S. Collins
Director, National Human Genome Research Institute
Before the
House Subcommittee on Labor, Health and Human Services
Education and Related Agencies
[house.gov]
March 12, 1998
Mr. Chairman and Members of the Committee: I am pleased to present the President's budget request for the National Human Genome Research Institute (NHGRI) for Fiscal Year 1999, a sum of $236,996,000, an increase of 10.4 percent over the FY 1998 comparable appropriation. Including the estimated allocation for AIDS, total support proposed for NHGRI is $240,134,000. Funds for the NHGRI efforts in AIDS research are included in the Office of AIDS Research budget request.
Gene Discovery
This is my fifth opportunity to appear before this Committee. As Director of NHGRI, I am proud of its leadership in the U.S. Human Genome Project and its cutting-edge research program on the genetic analysis of disease. Now at the half-way mark, progress in the 15-year Human Genome Project, its impact on health research and even public policy has surpassed the most ambitious expectations.
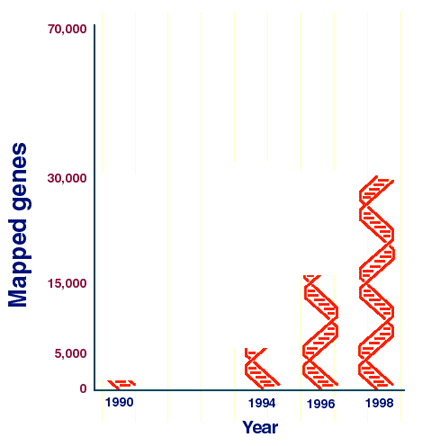
With Human Genome Project tools, it is possible to track down a disease-related gene even when nothing is known about the biochemical problems of the disease or how the gene works. This technique, based on identifying the position of a gene in the chromosome and then isolating it, was successfully used for the first time in 1986. Now, the increasing detail and quality of genome maps have reduced the time it takes to find a disease gene from years, to months, to weeks, to sometimes just days, and scientists are using the tools to discover dozens of disease genes each year. Increasingly, gene hunters are combining positional cloning techniques with a new "gene map" to make gene finding even easier and quicker. Constructed largely by scientists at NHGRI-supported research centers and the National Library of Medicine, the map has doubled in its detail since the first version was released two years ago. It now contains 30,011 gene tags, which may represent nearly half of all human genes (Chart 1). Disease-gene hunters now have about a 50 percent chance they will find an already characterized gene waiting for them in the chromosomal neighborhood they know is involved in a disease.
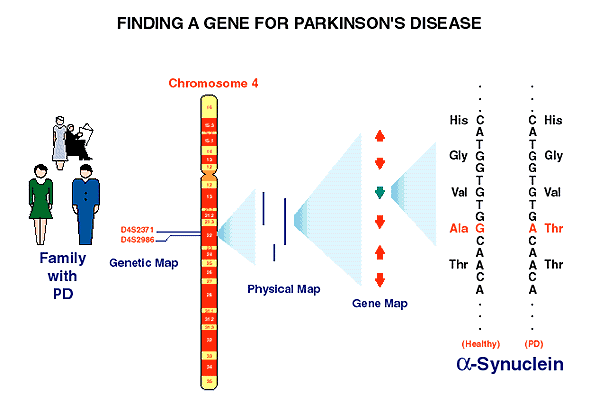
The isolation of a gene for Parkinson's disease (PD) last year demonstrated the power of this new discovery method and showed conclusively that changes in DNA can cause PD in some families. Only two years ago, the National Institute of Neurological Disorders and Stroke held a workshop to explore using genetic approaches to understand PD. A team led by scientists in NHGRI's Division of Intramural Research (DIR) began large-scale genetic analysis of DNA from members of a large Italian family containing almost 600 people, more than 60 of whom have been diagnosed with Parkinson's. In nine days, NHGRI gene hunters mapped the gene to a region of chromosome 4, which contained approximately 100 genes. One of the genes in that interval had already been identified on the gene map and was known to encode a protein called alpha-synuclein (chart 2). That gene was an excellent candidate for a Parkinson's disease gene because earlier research had already shown the protein builds up in brain cells of people with Alzheimer's disease, and people with PD have similar deposits. In just a few months, the researchers showed conclusively that an altered alpha-synuclein gene caused Parkinson's in the study families. Many have hailed this as the most significant advance in Parkinson's disease research in 30 years. Just ten days ago, a German research team used genome mapping tools to identify a new region on chromosome 2 that also appears to contain a gene that predisposes to Parkinson's. Some of the genes already known to exist in that region are providing excellent candidates for identification of the actual gene.
At NHGRI, intramural scientists and their colleagues have made tremendous progress this year in identifying the genetic components of some common cancers. These include identification of an altered gene that can cause multiple benign endocrine tumors and a type of pancreatic cancer - a syndrome called multiple endocrine neoplasia type 1 (MEN1), and a gene, called AIB1, linked to the growth and progression of breast cancer. Division of Intramural Research (DIR) scientists and their collaborators this past year isolated a gene underlying the neurological disorder Niemann-Pick Type C (NPC), and other genes involved in rare diseases that shed light on normal cell function. A newly discovered gene called JAG1, for example, was shown to cause the rare childhood disease Alagille syndrome. JAG1 has already been studied in the fruit fly and may help explain how incorrect cell signals during development can result in a wide range of birth defects.
Human Genome Project Progress
The ultimate goal of the international Human Genome Project is to spell out, letter by letter, all 3 billion bases in the human genome by the year 2005. Since the start of the Human Genome Project, scientists have been experimenting with whole-genome sequencing methods on smaller, less complex micro-organisms. This past year, for example, NHGRI-supported scientists at the University of Wisconsin-Madison [genetics.wisc.edu] published the full DNA sequence of the bacterium E. coli -- probably the most studied simple organism in all of science and the foundation of the biotechnology industry. Researchers now have access to the organism's genetics in their entirety -- all 4,403 genes nestled among 4,639,221 base pairs of DNA, which gives them a powerful new tool to observe complete cellular systems, including metabolism, the regulation of gene activity, substance transport and cell division.
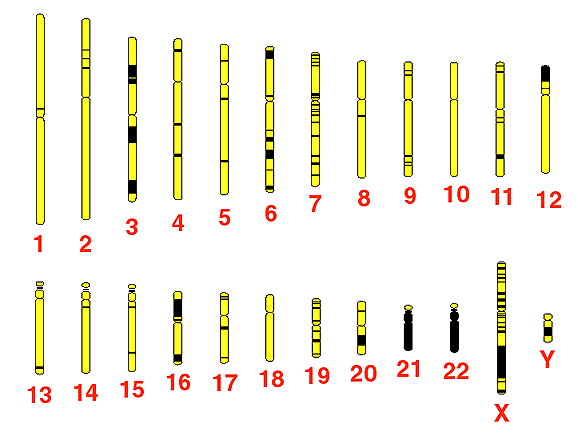
NHGRI began pilot projects in 1996 to test strategies and technologies for full-scale sequencing of the human genome, which contains about 1,000 times more DNA than E.coli does. Already, investigators have deposited almost 40 million bases of high-quality human DNA sequence in GenBank [ncbi.nlm.nih.gov]. NHGRI grantees expect to produce 60 percent or more of the total human DNA sequence (1.8 billion base pairs), with the remainder to be contributed by other Human Genome Project partners in the United States and abroad.(chart 3). To complete its portion, NHGRI recently announced a $70 million initiative to establish a coordinated network of laboratories. Those laboratories are expected to produce an average of 300 to 500 million bases of finished, accurate human sequence per year by 2003. The sequence produced must have four characteristics -- the "4 A's" of the Human Genome Project: 1) the sequence must be accurate, that is, the DNA spellings must be correct. The sequencing network will include quality assurance labs to ensure accuracy of 99.99 percent or better; 2) Large-scale sequencing relies on the accurate assembly of smaller lengths of sequenced DNA into longer, genomic-scale pieces, so DNA coming from the NHGRI network will be assembled into long pieces that reflect the original genomic DNA; 3) Because human DNA sequence must also be affordable, a portion of the network will focus on technology development to reduce cost as must as possible; 4) Finally, high-quality, finished human DNA sequence should be available to the entire research community, so NHGRI has introduced policies to make sequence from the network accessible within 24 hours through public databases.
The Human Genome Project is unique among most biological research efforts in its establishment of specific, goal-oriented research plans. The current plan expires in October, 1998, so NHGRI is in the midst of establishing a research plan to cover the next five years. The new plan will include working toward completing the first full human DNA sequence, developing faster sequencing technologies for the future, studying sequence variation and its relationship to disease, identifying and analyzing the function of genes in the human genome, and continued studies of the ethical, legal and social implications (ELSI) of new genetic technologies.
Complex Diseases
Straightforward rules of inheritance govern disease traits resulting from changes in a single gene, and it is now easier and faster than ever to track down and isolate such disease genes. But the inheritance of most common disorders -- diabetes, heart disease and most common cancers that result from the interplay of environment, lifestyle and small effects of many genes -- is much more complex and requires even more powerful tools.
NHGRI has just launched a new initiative to help speed the breakthroughs in complex disease research -- a major new goal for the Human Genome Project. Most complex diseases result from the combined effects of several relatively weak genetic contributions, and finding disease genes that have such weak effects has been painstakingly difficult. The search for such genes would be aided enormously by a thorough cataloging of the different versions of a whole list of genetic sites that occur in the human population. With such a catalog, scientists could begin to define which genetic differences are associated with a propensity for a specific disease and could help explain why, although we all carry the same genes, some individuals, families and even ethnic groups appear to be more likely than others to develop certain diseases. To facilitate these studies, NHGRI is leading an NIH-wide effort along with 17 other institutes to develop a very high-density map of slight-but-telling differences in the DNA code called "snips" (for single nucleotide polymorphisms or SNPs).
Another NHGRI initiative is contributing to the analysis of complex disease. The Center for Inherited Disease Research (CIDR), located on the Bayview campus of Johns Hopkins University, provides high-throughput genotyping services, study design advice, and sophisticated database assistance to research teams attempting to identify genetic locations and variations involved in complex diseases. A joint effort by eight NIH institutes, with the NHGRI serving as the lead, CIDR has just completed its first genotyping project for genomic changes associated with manic-depressive disorder. A second project, a study of deafness, has begun.
NHGRI is also involved in whole-genome studies to identify mutations that lead to adult onset diabetes mellitus and prostate cancer to understand and help remedy the disproportionately high rates of those diseases among African Americans. These studies are supported by the NIH Office of Research on Minority Health and organized by an NHGRI-Howard University collaborative center for studying diseases that disproportionately affect minority individuals.
Technology Development
Studies of genetic variation in large populations, complex diseases, and the simultaneous activity of multiple genes will rely on development of new technologies for large-scale genomic analysis. Such studies are already successfully using the marriage of semiconductors and DNA in so-called DNA chips. One version, which originated from an NHGRI grant to a scientist at a small California company, was recently featured on the cover of Fortune magazine and in this year's State of Union address. That chip consists of a thin slice of silicon about the size of a postage stamp upon which threads of DNA, whose spellings are already known, are arrayed. Such DNA chips can be used for a broad range of studies, including identifying DNA changes that lead to disease. This would be particularly useful, along with family histories and data from large population studies, for establishing an individual's risk of developing common, but hard-to-treat disorders like cancer, heart disease, diabetes, or psychiatric disorders, where multiple genetic alterations contribute, but each on a small scale. A baseline genome scan could give patients and health care providers helpful information about an individual's disease risk profile and point to which prevention strategies -- when available -- should be put into place. Eventually, the chips may even be used to identify which patients are genetically most likely to respond to specific therapies. Diseases may be subclassified by their underlying genetic configuration rather than by physical symptoms. Administering drugs targeted only to that particular genetic subtype could maximize efficiency, minimize side effects, and reduce treatment time wasted on ineffective therapies. The pharmaceutical industry is eagerly awaiting advances in DNA chip technology, and a catalog of human DNA variations, to incorporate into their research and development programs for improved treatment strategies.
Ethical, Legal and Social Implications (ELSI)
These dramatic new abilities to read large amounts of genetic information have important implications for the privacy and fair use of that information. The Human Genome Project is unique among research programs in its commitment to address the ethical, legal and social implications (See the Ethical, Legal and Social Implications Research Program) of its technologies side-by-side with the scientific agenda. NHGRI's ELSI research and policy development programs have spearheaded policy movement on a number of complex issues by supporting rigorous scholarly research and through development of options for state and federal policy makers.
One of the most active ELSI areas has been policy development related to the privacy and fair use of genetic information, particularly in health insurance, employment and medical research. This past year, President Clinton announced his support for a comprehensive legislative solution to the problem of genetic discrimination in health insurance, based on recommendations the ELSI Working Group and the National Action Plan on Breast Cancer (NAPBC) developed in a workshop three years ago. Protection gaps in The Health Insurance Portability and Accountability Act, which took a significant step toward preventing genetic discrimination in health insurance, can now be closed by legislation that would expand those protections to the individual insurance market. In January, Vice President Gore announced Administration support for federal legislation to prohibit genetic discrimination in the workplace. Again, the Administration's recommendations are based on the recommendations of the NAPBC-ELSI Working Group, which held a workshop two years ago to address the use of genetic information in employer-provided insurance, in discrimination in hiring and promotions, and privacy of that information. NHGRI staff worked closely with staff from the Office of the Secretary of Health and Human Services, the Department of Labor, and the Department of Justice to present these options to the White House. NHGRI and NAPBC recently met for a third time to address problems related to protecting the privacy of genetic information in the medical record. Workshop participants identified areas where new or modified policies or practices might enhance privacy protections without discouraging crucial areas of research. The group is developing a set of principles for researchers, research institutions, state and federal agencies, and policy makers to consider in formulating privacy protections for research information.
Other significant steps have been taken to ensure the responsible integration of genetic tests into clinical practice. The Task Force on Genetic Testing recently concluded its two-year analysis of genetic testing in the United States and published a report that contains recommendations for federal agencies, testing laboratories, and health professionals. The Cancer Genetics Studies Consortium, supported by NHGRI and several other NIH institutes, has published in medical journals their recommendations for follow-up care of individuals with specific cancer-causing gene mutations. Another product from the group describes informed consent for genetic testing for susceptibility to adult-onset cancer.
Recognizing the rapid pace of human genetics research and its impact on an increasing number of medical disciplines, the American Medical Association and the American Nurses Association, in collaboration with NHGRI staff, founded the National Coalition for Health Professional Education in Genetics. The coalition's mission is to ensure that our nation's health care providers have the knowledge, skills and resources to integrate responsibly new genetic knowledge into health care. Approximately 100 organizations representing health care professionals, consumer groups, industry, genetics professional organizations, and government agencies participated in the coalition's first meeting this past year. The new National Foundation for Biomedical Research, a government-created foundation "dedicated to creating public-private partnerships to promote the mission of the National Institutes of Health," is considering serving as the coalition's administrative manager.
As the Human Genome Project approaches the half-way mark, the contributions it has made to understanding human diseases have been gratifying and remarkable. In the years ahead, the full DNA sequence of the human will give us unprecedented opportunities to observe and understand the literal thread of life. There is now no question that genome maps, sequence and analytical tools will provide a robust technological infrastructure for biomedical research well into the next century.
The activities of the NHGRI are covered within the NIH-wide Annual Performance Plan required under the Government Performance and Results Act (GPRA). The FY 1999 performance goals and measures for NIH are detailed in this performance plan and are linked to both the budget and the HHS, GPRA Strategic Plan which was transmitted to Congress on September 30, 1997. NIH's performance targets in the plan are partially a function of resource levels requested in the President's Budget and could change based upon final Congressional appropriations action. NIH looks forward to Congress' feedback on the usefulness of its performance plan, as well as to working with Congress on achieving the NIH goals laid out in this plan. My colleagues and I will be happy to respond to any questions you may have.
Chart 1. The Gene Map of the Human Genome is being updated to contain 30,011 gene tags, representing nearly half the genes in the human genome. The number of mapped genes has doubled every two years since 1994.
 Return to the Statement Return to the Statement
Chart 2. Powerful gene-discovery methods helped to find the location of a gene for Parkinson's disease in just nine days. A gene map then gave researchers a likely candidate gene, which they soon confirmed was altered in family members with Parkinson's disease.
 Return to the Statement Return to the Statement
Chart 3. The human genome contains 3 billion DNA base pairs packaged on 24 different chromosomes. The first years of the Human Genome Project were devoted mostly to developing maps of the chromosomes, but now sequencing their DNA has begun in earnest. Sequencing has already begun in the areas marked in black, and by the year 2005, scientists will have determined the exact order of every base pair.
 Return to the Statement Return to the Statement
Last Reviewed: June 2006
|
 |

