|
Copyright © 2008, Eli Lilly and Company and the National Institutes of Health Chemical Genomics Center. All Rights Reserved. For more information, please review the Privacy Policy and Site Usage and Agreement.
Absolute EC50: The molar concentration of a substance that increases the measured activity in an agonist assay to 50% of the range of activity expressed relative to maximum and minimum controls.
Absolute IC50: The molar concentration of an inhibitor required to block a given response by 50%, half way between the maximum and minimum controls if the response is expressed relative to the maximum and minimum controls.
Accuracy: A measure of the closeness of agreement of a measured test result obtained by the analytical method to its theoretical true (or accepted reference) value. This is relevant only for calibration-curve based applications where a purified or relative reference standard material is available to quantify the analyte levels in test samples.
Accuracy Profile: A plot of the mean percent recovery (or percent relative error) and its confidence interval versus the concentration of the spiked standards (validation or QC samples). It is used to judge the quality of a calibration-curve based assay in terms of total error (bias plus variance).
Assays/Methods:
Assay-Biochemical: Biological measurements performed with purified or crude biochemical reagents.
Assay-Cell-based: Biological measurements performed in which at least one of the reagents consists of a population of live cells.
Assay Design: see Multivariate (Factorial Experiments), aka, Experimental Design.
Assay-In vitro: From the Latin meaning ‘in glass’. Any assay (biochemical or cell-based) conducted in a synthetic container (e.g. microtiter plate, microfluidic cell).
Assay-In vivo: From the Latin meaning ‘in life’. Typically used for assays conducted in living animals (e.g. mice, rats, etc.) with the exception of microorganisms (e.g. yeast, bacteria or C. elegans).
Assay Platform: Technology used to measure response or output. (e.g. Fluorescence polarization or Radiometric counting).
Assay-Phenotypic: An assay where the measured signal corresponds to a complex response such as cell survival, proliferation, localization of a protein, nuclear translocation etc. The molecular target is not assumed.
Assay-Primary: The first assay performed in a testing scheme to identify biologically active chemical entities in a screening mode.
Assay-Secondary: Assays that follow the primary assays to confirm the biological activity of chemical entities identified in the primary assays. This can also include selectivity and specificity assays.
Assay-Selectivity/Specificity: Assays employed to elucidate the specificity of biologically active chemical entities towards a set of closely related disease targets.
Assay Validation: see Validation
Assay-Separation: Prior to detection a physical separation of at least one component from the assay is performed. (Standard ELISA, filtration, HPLC etc.)
Assay-non-separation: Any assay where a physical separation is not required prior to detection.
Assay-Target based: An assay where the measured response can be linked to a known set of biological reagents such as a purified enzyme, domain or a reporter gene.
Biological Target: A macromolecule or a set of macromolecules in a biochemical pathway that is responsible for the disease pathology.
Bottom: The lower asymptote of a logarithmically derived curve. The Bottom value can be determined with real values or predicted using the logarithm applied to the result data set.
Calibration Curve: Also called “standard curve”, a calibration curve is a regression of the assay response on the known concentrations in “standard” samples. It is a model that fits data from standards and is used for calculating (calibrating) values of unknown test samples. For example, measurement of protein/biomarker expression levels of various compounds from in-vitro and in-vivo samples.
Central Composite Designs: A type of multi-factor experiment that is used to optimize the most important factors in an assay (usually 3 to 5 factors).
Classification & Regression Tree Models: A set of statistical methods in which observations are classified into groups based on a set of predictor variables and their relationship with a response variable. These models can be used for multivariate correlation analysis.
Cluster Analysis: A set of statistical methods in which objects (e.g., compounds) are divided into groups such that objects within a group are similar across a set of 2 or more variables.
Concordance Correlation Coefficient: A measure of agreement between two variables, i.e., how closely the paired values match. That is, it measures the degree of closeness of the data to the agreement line (Y=X line). Since the Pearson’s correlation measures the degree of departure of the data from the best straight line, which can be considerably different from the agreement line, the concordance correlation coefficient is more stringent than the Pearson’s correlation.
Control Compound: A compound that is routinely run in the same manner as the test compounds in every run of the assay. This term does not refer to the plate controls used to define the maximum and minimum responses, and they may or may not be a “literature standard” or “reference” compound.
CRC: Concentration-response curve mode. The mode to describe an assay performed with multiple concentrations of a given test substance, which might then render a logarithmically-derived graph curve.
Dynamic Range: It is the interval between the upper and lower concentration of the analyte in the sample for which the assay has been demonstrated to have acceptable level of accuracy, precision, linearity, etc.
EC50: The effective concentration of an agonist, which produces 50% of the maximum possible response for that agonist.
Emin: The maximum activity of an antagonist test substance relative to a reference agonist. This is obtained by first generating a fitted top from a %Inhibition curve and then converting that to the corresponding %Stimulation of the reference agonist curve. The E-min value for antagonist mode should equal the relative efficacy for agonist mode for competitive inhibitors.
Factor: An assay variable that can be changed by the user. Examples include the amount of a reagent, incubation time, buffer type, etc.
False Positive: A hit where the signal modulation is not related to the targeted activity. The sources of false positives include, random or systematic errors in liquid handling, spectrophotometric or fluorescence interference of the assay signal by chemical compounds, reagent instability etc. It is important to note that false positives can be reproducible when they are not related to random errors (as in the case of compound interference).
Fold Activity: The ratio of biological activity in the presence of an exogenous substance to that in its absence. It is the test compound’s observed response (raw data value) divided by the median of the same plate’s Min wells. This result type is used exclusively with single point assays. If the value is greater than 1, the test compound is likely an agonist. If the calculated value is less than 1, the test compound could be an inverse agonist.
Fold Activity Max: The maximum observed Fold Activity among the concentrations included in a concentration response curve. It is the test compound’s observed response (raw data value) divided by the median of the same plate’s Min wells. If the value is greater than 1, the test compound is likely an agonist. If the calculated value is less than 1, the test compound could be an inverse agonist.
Four Parameter Logistic Model: A non-linear regression model commonly used for fitting dose-response and concentration-response data. The four parameters are Minimum (response at zero dose), Maximum (response at infinite dose), Relative EC50 (or IC50, ED50, etc.) and Slope. The 4PL model can be written in several mathematically equivalent versions. Two popular versions are given below.
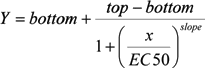
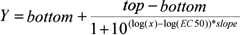
Fractional Factorial Experiments: A type of multi-factor experiment in which only a subset of factor level combinations is tested. These experiments are very efficient for screening a large number of factors prior to optimizing the most important factors.
Generalized Additive Models: Statistical models in which more general (e.g., nonlinear) relationships between variables can be examined. These models can be used for multivariate correlation analysis.
High Throughput Screening (HTS): Greater than 100,000 compounds screened per screen.
Homogeneous Assay: All assay components exist in solution phase at the time of detection (e.g. none of the components are in beads or cells). Technically no component scatters light.
Heterogeneous Assay: One or more assay components are present in solid phase at time detection. (e.g.: SPA, cells or IMAP).
Hill Coefficient: Derived slope a three or four parameter logistic curve fit. Should not be fixed to any given value without consultation with a statistician. It should not be a negative value except for inverse agonist assays.
Inhibition: Reduction of a predefined stimulus. Unit of Measure is always % when normalized to the dynamic range of the assay.
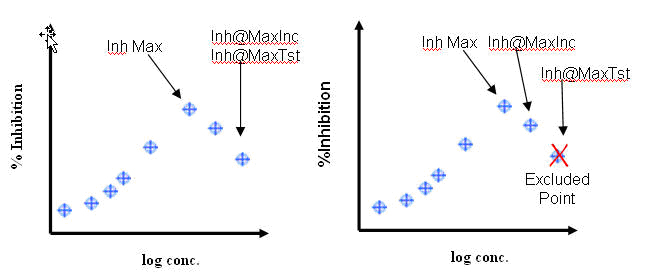
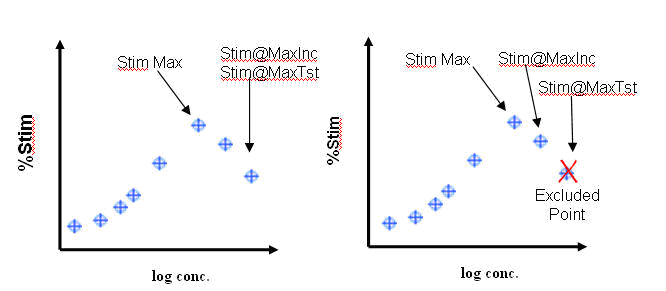
Inhibition at Max Included Concentration: Inhibition observed at the highest included (i.e. not excluded) concentration of a substance tested in a concentration response mode method version regardless of whether it was included in the parametric fit to produce derived results. (see Illustration below)
Inhibition at Max Tested Concentration: Inhibition observed at the maximum concentration of a substance tested in a concentration response mode method version regardless of whether it was included in the parametric fit to produce derived results. (see Illustration below)
Inhibition Max: Maximum inhibition produced by any concentration that was included for the application of a curve fit algorithm (see Illustration below)
Inverse Agonist: When an inverse agonist binds to a receptor, it stabilizes the inactive form of the receptor, shifts the equilibrium toward that state and produces a response opposite to that produced by an agonist in the biological system under investigation. These substances possess negative intrinsic activity.
Least Squares (Pearson’s) Correlation Coefficient: A measure of linear correlation between two variables.
LsA (Limits of Agreement): These are statistical limits that define the region that contains 95% of all potency ratios.
Mean Ratio (MR): The average ratio of potencies between the two runs.
Multivariate (Factorial) Experiments (Experimental Design): A system of experimentation for optimizing assays in which multiple factors are varied simultaneously in such a way that the effect of each factor can still is determined. In addition, one can also measure interactions between factors and use this information to more efficiently optimize an assay.
Multiple Linear Regression: A statistical method where the response variable is a linear function of several predictor variables. This can be used for multivariate correlation analysis.
Multivariate Correlation Analysis: A statistical analysis method where correlative relationships between 3 or more variables are examined.
Nonlinear Regression: Statistical methodology for fitting models that is nonlinear in their parameters, for example, the four-parameter logistic model.
One Factor at a Time Experiments: A series of experiments in which one factor is changed at a time. Once the “best” condition for one factor is found, it is fixed at that setting for subsequent experiments. This approach to assay optimization will not find the optimum conditions if at least one factor interacts with another, i.e., the best level of one factor depends on the levels of another factor.
Overall MSD (Minimum Significant Difference): The minimum difference in efficacies of two compounds evaluated in different runs that is statistically significant, i.e. that should be considered a real change in efficacy. The Overall MSD is defined for a single run of each compound.
Overall MSR (Minimum Significant Ratio): The minimum ratio in potencies of two compounds evaluated in different runs that is statistically significant, i.e. that should be considered real change in potency. The Overall MSR is defined for a single run of each compound.
Optimization: The process of developing an assay (prior to validation) wherein the variables affecting the assay are elucidated (e.g., Antibody concentration, incubation time, wash cycles, etc.). This process is ideally carried out using a multi-variate factorial approach where the inter-dependence between multiple variables/parameters can be taken into account.
Orphan Receptor: A biological target that has a primary sequence suggesting it is a member of one of the super families of biological targets; however, no ligand for this “receptor” has been identified. Generally, it is the aim of the research effort to identify ligands for this “orphan” so that a protocol for a validated assay can be created.
Percent Recovery: The calibrated value of a standard or validation sample divided by its expected value (known concentration), expressed as a percentage.
Plate Format: Microtiter plate well density (e.g., 96-, 384- or 1536-well) and plate composition (e.g., clear bottom black or clear bottom polystyrene, etc.)
Potentiation: Many assays involve the addition of one or more concentrations of a test substance in the presence of a fixed concentration of the known active substance called the Reference Agonist. In this mode, if an increased stimulus is observed the test compound is deemed a potentiator. Potentiation is the response produced by the combination of substances minus the response produced by the specific concentration of Reference Agonist alone.
Precision: A quantitative measure (usually expressed as standard deviation, coefficient of variation, MSR) of the random variation between a series of measurements from multiple sampling of the same homogenous sample under the prescribed conditions.
Precision Profile: A plot of the variability of calibrated values (expressed as a CV) versus concentration of standard. It is used to judge the quality of a calibration curve in terms of the variability in the calibrated values. It also determines the working range of a calibration curve.
Production MSD (Minimum Significant Difference): The minimum difference in efficacies of two compounds evaluated in different runs that is statistically significant, taking into account the number of runs routinely applied to all compounds in the assay. For example, if all compounds are routinely tested twice on separate days then the average of both runs will have greater precision than each individual run, and the Production MSD reflects this increased precision.
Production MSR (Minimum Significant Ratio): The minimum ratio in potencies of two compounds evaluated in different runs that is statistically significant, taking into account the number of runs routinely applied to all compounds in the assay. For example, if all compounds are routinely tested twice on separate days then the average of both runs will have greater precision than each individual run, and the Production MSR reflects this increased precision.
Quantitative Biology: A set of skills that is essential for the design, optimization and validation of reproducible and robust assays/methods to establish the pharmacological profiles of biologically active chemical entities. The practice of quantitative biology requires the understanding of how cellular, biochemical and pharmacological principles can be integrated with analytical and automation technologies, employing appropriate statistical data analysis and information technology tools.
Relative AUC: Defined as the ratio of the area under the fitted concentration-response curve for the test compound to the area under the fitted concentration-response curve for the reference compound.
Relative EC50: Relative EC50; the molar concentration of a substance that stimulates 50% of the curve (Top – Bottom) for that particular substance. It can also be described as the concentration at which the inflection point is determined, whether it’s from a three- or four-parameter logistic fit.
Relative EC50 Inv: The Relative EC50 of an inverse agonist.
Relative Efficacy: The maximum activity of a test substance relative to a standard positive control agonist. The result is expressed as percent from the following formula: 100 x Fitted Top of the test substance divided by the Fitted Top of an Agonist control. The agonist control should have a four parameter curve fit with defined lower and upper asymptotes but can have the Bottom fixed to zero in certain cases. The test compounds should have a four parameter curve fit but can have a three parameter fit with the bottom fixed to zero if the data warrants it.
Relative Efficacy Inv: The maximum activity of a test substance relative to a standard positive control inverse agonist. The result is expressed as percent from the following formula: 100 x Fitted Top of the test substance divided by the Fitted Top of the Inverse Agonist control. The inverse agonist control should have a four parameter curve fit with defined lower and upper asymptotes but can have the Bottom fixed to zero in certain cases. The test compounds should have a four parameter curve fit but can have a three parameter fit with the bottom fixed to zero if the data warrants it.
Relative IC50: Relative IC50; the molar concentration of a substance that inhibits 50% of the curve (Top – Bottom) for that particular substance. It can also be described as the concentration at which the inflection point is determined, whether it’s from a three- or four-parameter logistic fit.
Relative Potentiator Efficacy: The fitted top of the potentiation curve minus the normalized response to the specific concentration of Reference Agonist alone divided by 100 minus the normalized response to the specific concentration of Reference Agonist alone.
Response Surface Analysis: A statistical analysis method that is used for central composite designs. A quadratic polynomial model is fit to the data in order to find the optimum conditions for an assay.
Repeatability: Repeatability is the precision of repeated measurements within the same analytical run under the same operating conditions over a short interval of time. It is also termed intra-assay or intra-batch precision.
Reproducibility (Run to Run): A general term to describe the precision of results generated from multiple runs of a compound (or any homogenous test sample) in an assay. An assay may lack reproducibility because of either high within-run or across-run variability, or because of systematic trend (drift) over time in the response. An assay that is reproducible across runs is one that has variation within acceptable limits and has no material systematic trends.
Reproducibility (Lab to Lab): Reproducibility across labs expresses the precision between laboratories. It is useful for assessing the “transferability” of an assay and/or the validity of comparing results from samples that are run in two or more laboratories.
Robustness/Ruggedness of the Assay: Robustness is a measure of the capacity of the assay to remain unaffected by small, but deliberate changes in method parameters and provides an indication of its reliability during normal run conditions.
Schild Kb: A measure of affinity for a competitive antagonist that is calculated using the ratios of equi-active concentrations of a full agonist (most typically EC50 concentrations are used) in the absence and presence of one or more concentrations of the antagonist. See pp. 335-339, Pharmacologic Analysis of Drug-Receptor Intercation, 3rd Ed. by Terry Kenakin.
Signal-to-background (S:B or S/B): The ratio between the mean max signal and the mean minimum or background signal. This definition has been useful for describing the dynamic range of the assay.
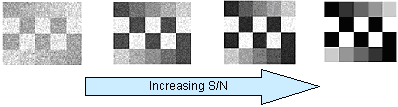
Signal to Noise Ratio (S:N or S/N): Defined as the mean signal (eg. Max or Min signal) divided by the standard deviation of this signal. This definition has been useful for describing the “signal strength” of an assay. The figure below illustrates how a two signals (black= background or Min and white=max) may appear as the “noise” in the system varies.
Signal Window: A measure of separation between max. and min. controls in an assay that accounts for the amount of variability in the assay. The formula is:
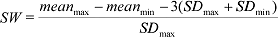
Simple Linear Regression: A statistical method for fitting a straight line to paired (X, Y) data.
Spearman’s Correlation Coefficient: A nonparametric measure of correlation between two variables. It is applied to ranked values of the data and is therefore robust to outliers in the data.
Specificity: The ability of the assay to determine unequivocally the analyte in the presence of components that may be expected to be present in the sample.
Spike: Addition of a known quantity of a specific reference material or positive control to a sample matrix for recovery studies.
Stephenson’s Kp: A measure of affinity for a partial agonist that is calculated through the comparison of equi-active concentrations of a full agonist in the absence and presence of a single concentration of the partial agonist. See pp. 284-286, Pharmacologic Analysis of Drug-Receptor Intercation, 3rd Ed. by Terry Kenakin.
Stimulation: Increase of a measured output. Unit of Measure is always % when normalized to the dynamic range of the assay (Min and Max control wells). Note that this calculation can generate percents much higher than 100.
Stimulation at Max Included: Stimulation observed at the highest included (i.e. not excluded) concentration of a substance tested in a concentration response mode method version regardless of whether it was included in the parametric fit to produce derived results. (See illustration below)
Stimulation at Max Tested: Stimulation observed at the maximum concentration of a substance tested in a concentration response mode method version regardless of whether it was included in the parametric fit to produce derived results. (See illustration below)
Stimulation Max: Maximum stimulation produced by any concentration that was included for the application of a curve fit algorithm (See illustration below)
Target Platform: A set of biochemically and biologically related targets implicated in disease pathologies. Examples include G-protein coupled receptors (GPCRs), nuclear hormone receptors (NHRs), Kinases, Proteases, Transporters, Ion channels and Chemokines.
Testing Flow Scheme: Stages of testing NCEs as it progresses from active to hit to lead to a clinical candidate. Tests include in-vitro assays, animal model tests, ADME assays biopharmaceutical and toxicological tests.
Test-retest experiment: An experiment in which a set of (usually) 20-30 compounds is tested in two independent runs of an assay. Its purpose is to estimate the MSR (for dose-response assays) or the MSD (for single-point assays or an efficacy measure in dose-response assays). This experiment will provide a reliable estimate of only the within-run MSR.
Top: The upper asymptote of a logarithmically derived curve. The Top value can be determined with real values or predicted using the logarithm applied to the result data set. The Unit of Measure is always %.
Ultra High Throughput Screening (uHTS): Greater than 500,000 compounds screened per screen.
Validation: Validation includes all the laboratory investigations that demonstrate that the performance characteristics of an assay are suitable and reliable for its intended analytical use. It describes in mathematical and quantifiable terms the performance characteristics of an assay.
Validation/QC Samples: Samples of standard material that are prepared independently of the standards used for a calibration curve. They are not used to fit the calibration curve, but they are calibrated against it to determine percent recovery.
Within-Run MSD (Minimum Significant Difference): The minimum difference in efficacies of two compounds evaluated in the same run that is statistically significant, i.e. that should be considered a real change in efficacy.
Within-Run MSR (Minimum Significant Ratio): The minimum ratio in potencies of two compounds evaluated in the same run that is statistically significant, i.e. that should be considered real change in potency.
Z’-Factor: Another measure of separation between maximum and minimum controls in an assay that accounts for the amount of variability in the assay. The formula is:
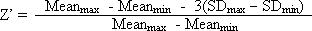
|