|
 Previous Event | Next Event Previous Event | Next Event 
1968: First Restriction Enzymes Described
 Several groups of researchers – including M. Meselson’s group at Harvard and H. O. Smith, K. W. Wilcox, and T. J. Kelley at Johns Hopkins – studied and characterized the first restriction nucleases, enzymes that revolutionized molecular biologists’ ability to manipulate DNA. Several groups of researchers – including M. Meselson’s group at Harvard and H. O. Smith, K. W. Wilcox, and T. J. Kelley at Johns Hopkins – studied and characterized the first restriction nucleases, enzymes that revolutionized molecular biologists’ ability to manipulate DNA.
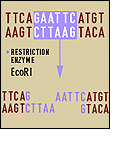
Restriction enzymes recognize and cut specific short sequences of DNA. They’re found in bacteria, which use the enzymes to digest invading DNA. The bacteria add methyl groups to their own DNA to protect them from digestion. Molecular biologists began using these enzymes, along with DNA polymerase and DNA ligase (an enzyme that sticks fragments of DNA together), in the early 1970s to cut, manipulate, and analyze pieces of DNA in a predictable and reproducible way. The enzymes became an important, early tool for mapping genomes.
More Information
The first restriction nuclease characterized was isolated from Haemophilus influenzae bacteria. The enzyme (HindII) cuts at a particular site within a specific sequence of six base pairs as follows. (Where nucleotides are shown in parenthesis, the enzyme recognizes either one of the two bases shown.) Other restriction nucleases cut at other specific sites within specific sequences.
5'–GT(T/C)(A/G)AC–3' HindII 5'–GT(T/C)–3' 5'–(A/G)AC – 3'
3'–CA(A/G)(T/C)TG–5' ----> 3'–CA(A/G)-5' + 3'–(T/C)TG – 5'
References:
Luria, S.E. The Recognition of DNA in Bacteria. Scientific American, 222:88-92. 1970. [PubMed]
Smith, H.O., Wilcox K.W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol, 51: 379-91. 1970. [PubMed]
Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature, 217 (134):1110-4. 1968. [PubMed]
 Previous Event | Next Event Previous Event | Next Event 
Last Reviewed: April 15, 2008
|

