|
InterProScan (IPRScan) is a tool that combines different protein signature recognition methods into one resource. IPRScan not only wraps the sequence analysis applications, it also performs a considerable amount of program outputs and data look-up from various databases. InterPro (IPR) integrates PROSITE, PRINTS, Pfam, ProDom, SMART, TIGRFAMs, PIR superfamily, SUPERFAMILY, Gene3D and PANTHER databases. Each member database in IPR uses different scanning applications. InterPro and Interproscan are developed and maintained at the EMBL-EBI. IPRScan on Biowulf is designed to be used for bulk sequence analysis such as hundreds or thousands of sequences. For small scale of sequence analysis, please use the EBI IPRScan website which can handle 1 sequence at a time. |
- Prepare the input sequence file by putting all the sequences of the same format in one file.
- Current settings for IPRScan on Biowulf.
- max amino acids for input sequence=3000
- max nucleic acids for input sequene=1000
- max length for the nucleotide input sequence=10000
- min length for the protein input sequence=5
- default minimum orf size for translation=50
- Most major sequence formats are acceptable, including Fasta, Genbank, EMBL, GCG and Swissprot. IPRScan reformats input sequences using the 'seqret' program in EMBOSS. For detailed list of acceptable sequence format, see the 'Input sequence formats' section in the EMBOSS seqret documentation.
- Do not mix nucleotide and amino acid sequences in a single file.
- Do not mix different formats of sequences in a single file.
- Sample input file:
>RS16_ECOLI MVTIRLARHGAKKRPFYQVVVADSRNARNGRFIERVGFFNPIASEKEEGTRLDLDRIAHW VGQGATISDRVAALIKEVNKAA >Q9RHD9 XPKLEEGVEGLVHVSEMDWTNKNIHPSKVVQVGDEVEVQVLDIDEERRRISLGIKQCKSN PWEDFSSQFNKGDRISGSIKSITDFGIFIGLDGGIDGLVHLSDISWNEVGEEAVRRFKKG DELETVILSVDPERERISLGIKQLEDDPFSNYASLHEKGSIVRGTVKEVDAKGAVISLGD DIEGILKASEISRDRVEDARNVLKEGEEVEAKIISIDRKSRVISLSVKSKDVDDEKDAMK ELRKQEVESAGPTTIGDLIRAQMENQG >Y902_MYCTU Q10560 PROBABLE SENSOR-LIKE HISTIDINE KINASE RV0902C (EC 2.7.3.-). MNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKERLDRRLDEAAGFA IPFVPRGLDEIPRSPNDQDALITVRRGNVIKSNSDITLPKLQDDYADTYVRGVRYRVRTV EIPGPEPTSVAVGATYDATVAETNNLHRRVLLICTFAIGAAAVFAWLLAAFAVRPFKQLA EQTRSIDAGDEAPRVEVHGASEAIEIAEAMRGMLQRIWNEQNRTKEALASARDFAAVSSH ELRTPLTAMRTNLEVLSTLDLPDDQRKEVLNDVIRTQSRIEATLSALERLAQGELSTSDD HVPVDITDLLDRAAHDAARIYPDLDVSLVPSPTCIIVGLPAGLRLAVDNAIANAVKHGGA
/usr/local/iprscan/bin/iprscan -cli -h
|
Mandatory options |
|
| -cli | use command-line interface. Required for Biowulf runs. |
| -i | Input sequence file. |
| -seqtype n | Required for nucleotide sequences |
|
Other options |
|
| -o outputfilename | The output file where to write results (optional), default is STDOUT which is /home/UserID (see table below). |
| -email <addr> | Submitter email address (Not required for Biowulf batch system). |
| -appl <name> | Application(s) to run (optional), default is all. Possible values (dependent on set-up): blastprodom fprintscan hmmpfam hmmpir hmmpanther hmmtigr hmmsmart superfamily gene3d scanregexp profilescan seg coils |
| -nocrc | Don't perform CRC64 check and rerun all searches even already exist in database which is unnecessary. The default is run without this flag. |
| -altjobs | Launch jobs alternatively, chunk after chunk. Default is off. |
| -seqtype <type> | Sequence type: n for DNA/RNA, p for protein (default). |
| -trlen <n> | Transcript length threshold (20-150). |
| -trtable <table> | Codon table number. |
| -goterms | Show GO terms if iprlookup option is also given. |
| -iprlookup | Switch on the InterPro lookup for results. |
| -format <format> | Output results format (raw, txt, html, xml(default), ebixml(EBI header on top of xml), gff) |
| -verbose | Print messages during run |
#!/bin/bash # #PBS -N YourScriptNameHere #PBS -m be #PBS -k oe /usr/local/iprscan/bin/iprscan -cli -i /data/maoj/iprscan/test248aa.seq -o /dev/null \ -format raw -goterms -iprlookup
- Run the following command on Biowulf:
<biowulf %> qsub -l nodes=1 YourScriptNameWithFullPath
- To check status of your job:
<biowulf %> qstat -u YourUserID
- More info about monitoring your jobs.
- Sequences in the input file will be split into chunks of 100. For example, 250 sequences will be split to 3 chunks.
Directory/File Example Notes /home/YourUserID/YourJobName.oJobNo
/home/YourUserID/YourJobName.eJobNo
/home/YourUserID/iprscan-xxx.oxxxxx
/home/YourUserID/iprscan-xxx.exxxxx/home/userID/growth.e1151902
/home/userID/growth.o1151902
/home/userID/iprscan-2008050.e1269981
/home/userID/iprscan-2008050.o1269981YourJobName is the name specified in the batch script file, beside "#PBS -N".
The JobNo is the number appears right after user submit the job.
If -o option is not given, the summary result not only will appear in 'merged.raw' file (see below) but also in this 'xxx.oJobNo' file. To stop the result from duplicating, include '-o /dev/null' in the command as appeared in sample script above.
/data/YourUserID/iprscan-yyyymmdd/ /data/userID/iprscan-20080301 Automatically created /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx//data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/Subdirectories created based on time stamp the job is submitted if multiple jobs are submitted in the same day. /data/UserID/pxxxxxxxx.res or .log or .fa /data/UserID/p1063121154651720535.fa
/data/UserID/p1063121154651720535.log
/data/UserID/p1063121154651720535.resThese files are temperatory and will be cleaned up automatically before job finishes. Do not touch or remove these files when the job is running. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/iprscan-
yyyymmdd-hhmmssxx.exitcode/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/
iprscan-20080301-11414247.exitcodeFile content should be '0' if job runs successfully. However, .exitcode files under all chunks should be double checked to confirm. User can go to each chunk_x directory and type 'more iprscan*.exitcode' to view all the exitcode files content at once. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/
iprscan-yyyymmdd-hhmmssxx.input/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/
iprscan-20080301-11414247.inputInput file with sequence format converted to fasta format /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/
iprscan-yyyymmdd-hhmmssxx.input.inx/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/
iprscan-20080301-11414247.input.inxBinary format of input sequences /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/
iprscan-yyyymmdd-hhmmssxx.params/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/
iprscan-20080301-11414247.paramschecksum summary of all the input sequences /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/
iprscan-yyyymmdd-hhmmssxx.seqs/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/
iprscan-20080301-11414247.seqsInput file with sequences of original format /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/merged.rawIn addition to merged.raw, it can also be xxx.html or xxx.xml or xxx.txt depend on the format user specified.
/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/merged.raw
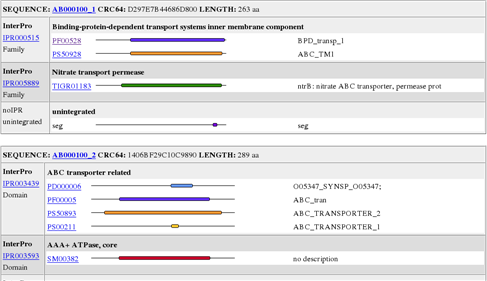
html output sample :
Output summary file of all chunks. The format can be merged.raw or html or xml or txt. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/chunk_x//data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/chunk_3Directories created for each chunk of sequences which contains output files for each of the 13 applications. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/chunk_x/
iprscan-yyyymmdd-hhmmssxx-APP-cnkX.
OUTPUTFILE/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/chunk_2/
iprscan-20080310-13280403-profilescan-cnk2.
exitcodeEach application generates 4 output files: .output; .output.inx; .errors; .exitcode. Check all the exitcode output file for each application in each chunk. The content should be '0' in all exitcode files for a successful run. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/chunk_x/
iprscan-yyyymmdd-hhmmssxx.nocrc/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/chunk_2/
iprscan-20080310-13280403.nocrc.nocrc file under each chunk_x directory contains the query sequences that do not have a known crc64 according to the match.xml file. Applications will only be launched against these sequences if -nocrc flag is not issued. /data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/chunk_x/
iprscan-yyyymmdd-hhmmssxx.xml/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/chunk_2/
iprscan-20080310-13280403.xml'xxx.xml' file under each chunk_x directory contains the default output result from the search for each chunk. Additional output format can be obtained by changing the command option from -format raw to -format html for example.
To view .html output file, type 'firefox YourFileName.html'
/data/YourUserID/iprscan-yyyymmdd/
iprscan-yyyymmdd-hhmmssxx/chunk_x/merged.raw/data/YourUserID/iprscan-20080301/
iprscan-20080301-11414247/chunk_2/merged.rawmerged.raw file under each chunk_x directory contains the output in raw format converted from .xml file.
Output files from iprscan runs are grouped under two directories: /home/userID & /data/userID/iprscan-xxxxx. Since the number of files can accumulate and fill up user's space fast, frequent cleanup by users themselves is highly recommended.
The main output file(s) such as merged.raw or xxx.html or xxx.xml or xxx.txt contains the summarized interesting output from all chunks in each run. The other files can be deleted after checking exitcode files in the chunk_x directories as described above. Sample cleanup commands:
% cd /data/user % mv iprscan-yymmdd/iprscan..../merged.raw . # or *.html or *.txt or *.xml % rm -r iprscan-yymmdd
For these benchmarks, an input file containing query sequences was submitted to batch system using qsub -l nodes=1,mem=2048 scriptName as in the example above. Jobs were run on ??? nodes with 2 GB of RAM.
- 3000 protein query sequences: 1 hour 32 minutes
- 1000 protein query sequences: 57 minutes
- 1000 nucleotide query sequences: 2 hours 50 minutes
- The InterPro database is updated within a week whenever a newer version is available.
- Database files can be accessed at /fdb/iprdb/data.new
Please contact the Helix Systems staff if you have questions.