eyeGENETM - Funding Trends and Supported Activities

The following tables and their representative graphs depict the funding allocation of the eyeGENETM initiative since its launch. The data show the dynamic evolution of eyeGENETM over its brief lifespan.
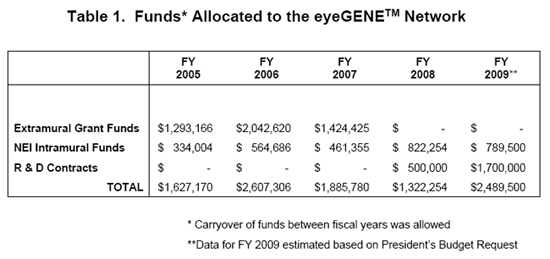
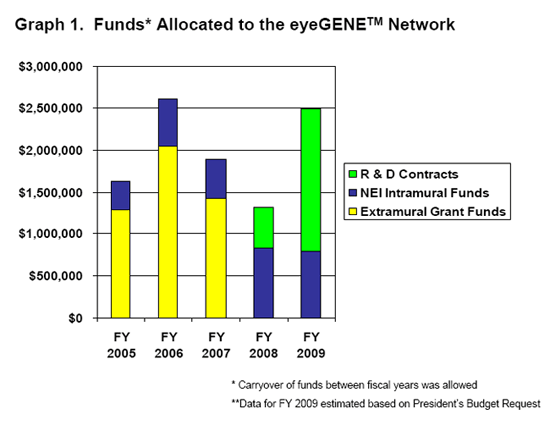
Representation of the funding allocation to the eyeGENETM Initiative broken down into the three major sources of funding: Extramural Grant Funds, NEI Intramural Program Funds and R & D Contracts.
The Extramural Grant Funds allocation represents Administrative Supplements awarded to researchers and clinicians who have ongoing NEI-supported projects and who have access to or are part of Clinical Laboratory Improvement Amendment of 1988 (CLIA)-approved facility to build the capability for molecular genetic testing for ocular diseases. This initiative was developed and implemented in response to a recommendation from the National Advisory Eye Council (NAEC) for the establishment of a National Ophthalmic Disease Genotyping Network.
The NEI Intramural Funds allocation represents eyeGENETM funds allocated to the NEI CLIA Diagnostic Laboratory and to the Ophthalmic Genetics Visual Function Branch (OGVFB), located within the NEI Intramural Research Program in Bethesda, MD.
The R & D Contracts line represents contracts awarded to offerors who furnish all the necessary services, qualified personnel, materials, equipment and facilities, not otherwise provided by the Government, as needed to be a full collaborating participant in the eyeGENETM Network. The Contracts mechanism is replacing the grant supplement mechanism as the primary means to support this initiative.
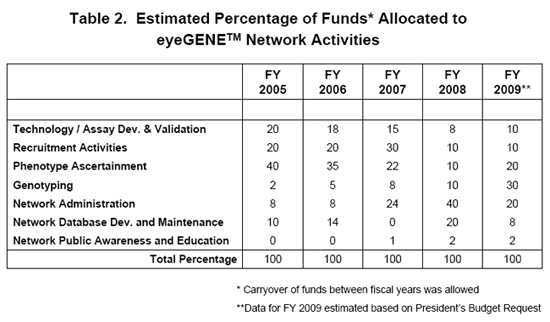
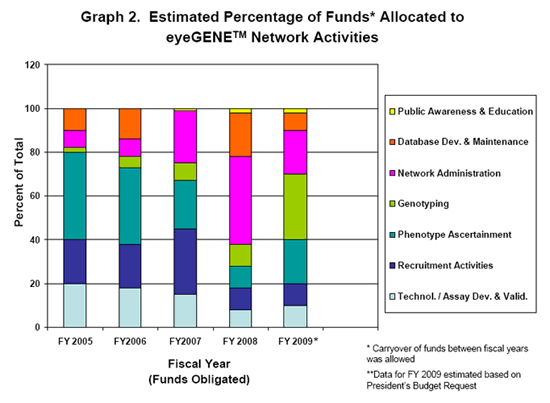
This national Network of collaborating research and CLIA laboratories fosters academic collegiality and cooperation across eye research disciplines. It also includes community eye health care providers who are not associated with an academic institution, i.e. private practice clinicians. The end result will be a more informed vision community with data sharing, a repository and database for future research efforts into genetic eye diseases paving the way for potential treatments.
The activities of the Network have been broken down into 7 broad categories. They are not all inclusive, as eyeGENETM is ever-evolving, but they represent a snapshot of activities performed by members of the Network.
The categories are as follows:
Technology & / or Assay Development
This category focuses on the development of genotyping microarrays for high-throughput, efficient, and affordable genetic screening of genetic eye diseases.
It also includes the development of new gene assays and validation techniques before a gene can be added to the eyeGENETM gene testing menu.
Recruitment Activities
Each participating member of the eyeGENETM Network has adapted their ongoing independent research protocols and patient consent forms and received Institutional Review Board (IRB) approval to participate in the Network. Members have agreed to follow NIH guidance regarding resource and data sharing, and policies regarding public access. As a result, these members can recruit patients with genetic eye disease into the Network.
Phenotype Ascertainment
Phenotyping is the cataloging of observable traits or characteristics of an individual. Family history of eye disease and pedigrees of eyeGENETM study participants are gathered. This involves the collection of data and samples, developing data intake and forms that capture information from detailed eye examinations.
Genotyping
Genotyping involves the identification of the genetic makeup of an individual, in this case, identifying a disease-causing mutation in an individual with genetic eye disease. Currently, eyeGENETM members can test for mutations in approximately 50 disease-causing genes.
Network Administration
The eyeGENETM Coordinating Center currently resides in the NEI Intramural Program as part of the activities of the Ophthalmic Genetics Visual Function Branch (OGVFB). The eyeGENETM Coordinating Center is responsible for receipt of participant samples, processing of blood to DNA, shipping of DNA to an eyeGENETM CLIA-Network laboratory for diagnostic testing. The Coordinating Center is responsible for eyeGENETM protocol amendments, verification that protocols and documentation / certifications at various participating Network institutions are renewed when appropriate. The Center coordinates various activities among the Network-associated organizations. The Center also maintains a call log and handles patient inquiries and assists clinicians / researchers from various organizations, as well as Network members in the use of the eyeGENETM online database. The Coordinating Center maintains the eyeGENETM database and performs security testing when database versions are updated. The Center also oversees quality assurance testing among the Network laboratories.
The Coordinating Center also interacts with an external advisory group which provides oversight and input into Network operations and direction.
Database Development and Maintenance
A database was developed especially for the eyeGENETM Initiative. The types of data captured in the eyeGENETM database consist of patient information, family information, clinical findings, medical images and other uploaded files, consent forms, genetic test results, and tracking data for the flow and processing of blood and DNA specimens. ALL of this data is protected so that only the referring eye health care provider and the Coordinating Center have access to this information. The CLIA laboratories performing the molecular diagnostic testing have access only to unique identifiers (unless they are the referring clinician).
The eyeGENETM database is a good example of the integration of health Information Technology and genetic information. Its use of dynamic phenotype content, for example, allows support for emerging new diseases and diagnostic tools, and an optimal balance of providing useful clinical data for researchers, minimal burden for clinicians, and compatibility with medical information standards. There are also advanced security and audit trails in the system to protect confidential information.
When the eyeGENETM repository hits a certain critical mass and is available to the vision community ALL of the data will be extracted into a de-identified view that will be available to researchers who apply for access and samples.
Public Awareness & Education
This activity includes meetings for the eyeGENETM Steering Committee and the Network participants. It also includes web materials and brochures for both the research / clinical community and patients with genetic eye diseases who are interested in eyeGENETM.