|
One man who drinks alcohol and smokes cigarettes lives to age 90 without getting liver or lung cancer; another man who smokes and drinks the same amount gets cancer at age 60.
One woman's breast cancer responds to chemotherapy, and her tumor shrinks; another woman's breast cancer shows no change after the same treatment.
How can these differences be explained?
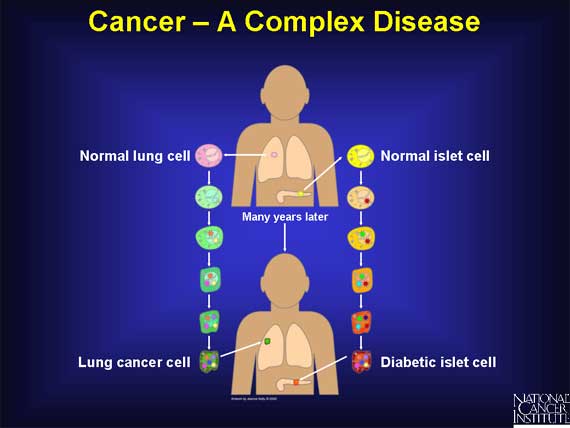
Cancer is a complex disease, like diabetes, heart disease, and kidney disease. All complex diseases arise from combinations of changes that occur in the same cell over a period of time. In cancer, these changes accumulate over many years. Only when a critical number of changes occur in the same cell does it finally become cancerous.
To complicate matters further, there are many different types of cancer, each displaying different combinations of changes. Even within a single type, such as lung cancer or colon cancer, clinicians can identify subtypes, each marked by a unique set of changes.
How can researchers hope to identify and study all the changes that occur in so many different cancers?
How can they explain why some people respond to treatment and not others?
How can they explain why some smokers get lung cancer, and others do not?
How can these differences be explained?
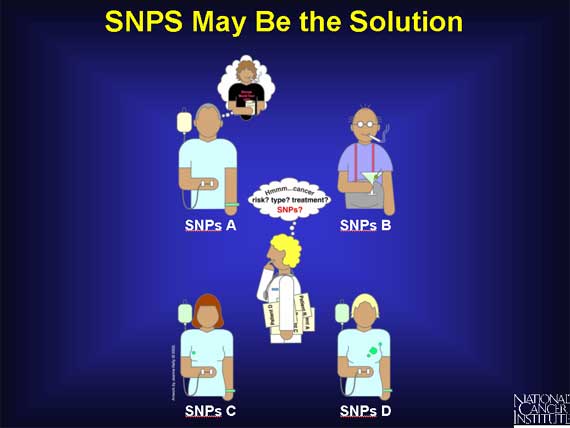
Scientists think that tiny variations in the human genome called Single Nucleotide Polymorphisms, or SNPs (snips) for short, can help them to answer these questions. They believe SNPs can help them catalogue the unique sets of changes involved in different cancers. They see SNPs as a potential tool to improve cancer diagnosis and treatment planning. They suspect that SNPs may play a role in the different responses to treatments seen among cancer patients. And they think that SNPs may also be involved in the different levels of individual cancer risk observed.
So what exactly are SNPs?
How are they involved in so many different aspects of health?
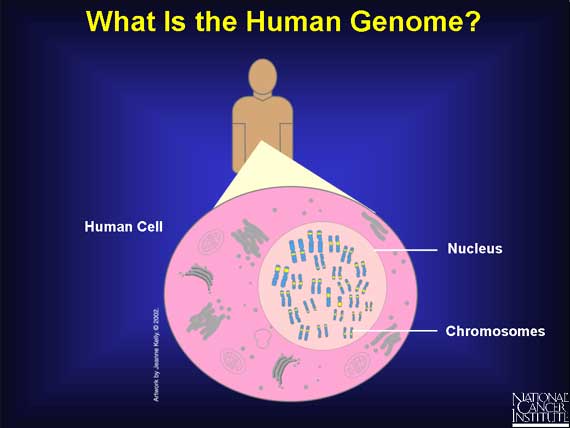
To answer these questions, one first needs to look at the human genome.
The human genome is the complete set of instructions for life as we know it. Except for red blood cells, which have no nucleus, the human genome is located in the nucleus of every cell in the body. There it is organized into 22 pairs of very large molecules called chromosomes and one pair of sex chromosomes.
Chromosomes are made of deoxyribonucleic acid (DNA).
DNA contains only four chemical bases or building blocks: Adenine, Thymine, Cytosine, and Guanine - called A, T, G, and C, for short. There are roughly 3.2 billion chemical bases (A, T, C, G) in the human genome.
Each DNA molecule is made up of two long complementary (related) strands, which are intertwined like a rope. This is the famous "double helix." Since A always pairs with T, and C with G, the order on one strand dictates the order on the other. This is called base pairing and enables the genome to make copies of itself.
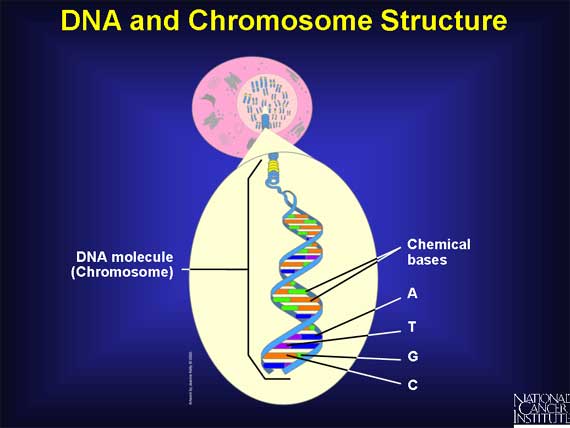
Only about 3 percent of the human genome is actually used as the set of instructions. These regions are called coding regions, and they are scattered throughout the chromosomes.
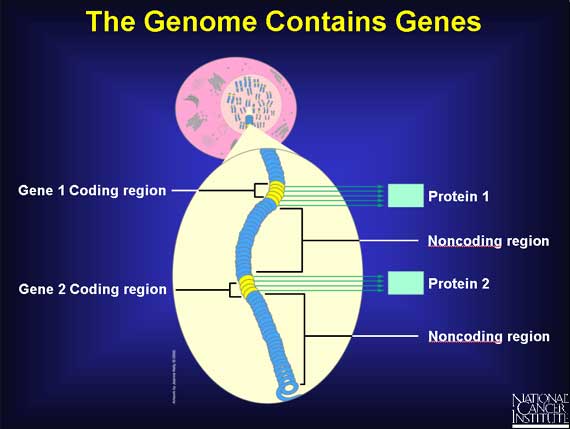
A coding region contains genes. A gene is a unique DNA sequence within a chromosome that ultimately directs the building of a specific protein with a specific function. Close to each gene is a "regulatory" sequence of DNA, which is able to turn the gene "on" or "off." There are at least 35,000 genes in the human genome, and there may be more.
Scientists can find no function for most of the remaining 97 percent of the genome - yet! These regions are called noncoding regions.
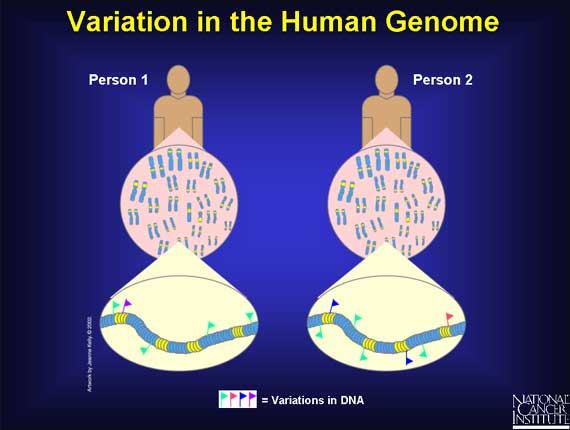
An amazing aspect of the human genome is that there is so little variation in the DNA sequence when the genome of one person is compared to that of another. Of the 3.2 billion bases, roughly 99.9 percent are the same between any two people. It is the variation in the remaining tiny fraction of the genome, 0.1 percent--roughly several million bases--that makes a person unique. This small amount of variation determines attributes such as how a person looks, or the diseases he or she develops.
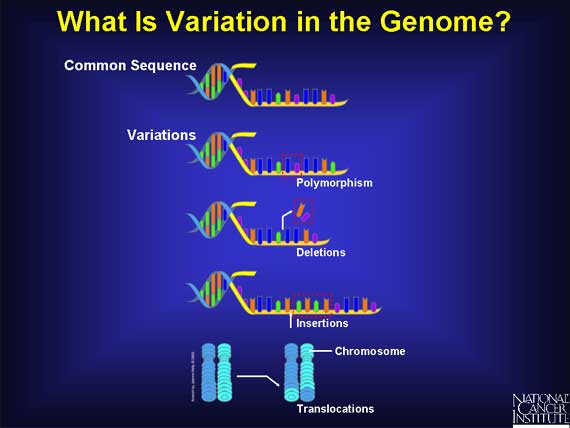
Variation occurs whenever the order of the bases in a DNA sequence changes. Variations can involve only one base or many bases. If the two strands of a chromosome are thought of as nucleotides threaded on a string, then, for example, a string can break and the order of the beads can vary. One or more nucleotides may be changed, added, or removed. In chromosomes, these changes are called polymorphisms, insertions, and deletions.
In addition to these changes, some DNA sequences called "repeats" like to insert extra copies of themselves several times. Chromosomes can also undergo more dramatic changes called translocations. These occur when an entire section of DNA on one chromosome switches places with a section on another.
Not all variations in the genome's DNA sequences have an effect. Among the variations that do cause effects, some are more serious than others. The outcome depends on two factors: where in the genome the change occurs, (i.e., in a noncoding region, coding, or regulatory region) and the exact nature of the change.
Most variations in the human genome have no known effect at all because they occur in noncoding regions of the DNA. In addition, there are some changes that do occur in coding and regulatory regions, yet they have no known effect. All these are silent variations.
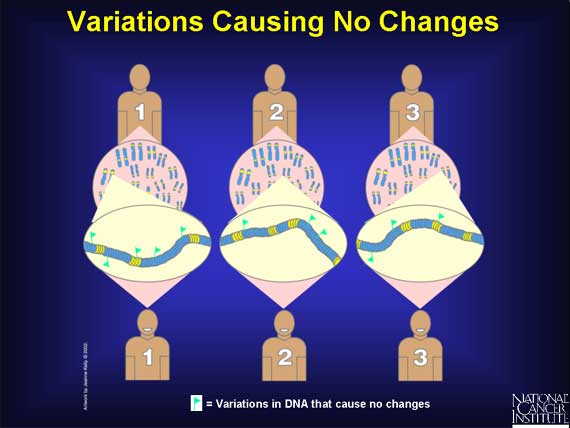
Some of the variations that occur in the coding and regulatory regions of genes have "harmless" effects. They can, for example, change the way a person "looks." Some people have blue eyes, others brown; some are tall, others short; and some faces are oval, others round.
Other variations in coding regions are harmless because they occur in regions of a gene that do not affect the function of the protein made.

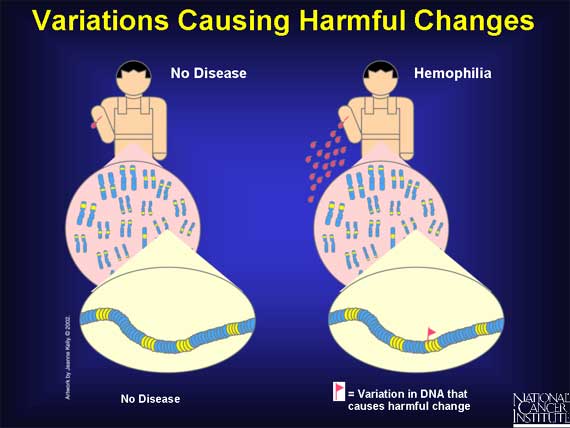
There are a group of variations in coding and regulatory regions that result in harmful effects. These are called mutations. They cause disease because changes in the genome's instructions alter the functions of important proteins that are needed for health. For example, diabetes, cancer, heart disease, Huntington's disease, and hemophilia all result from variations that cause harmful effects.
In a "simple" disease such as hemophilia, variation in one gene is sufficient to cause disease symptoms. By contrast, in a "complex" disease like cancer, symptoms are seen only after many variations have occurred in different genes in the same cell.
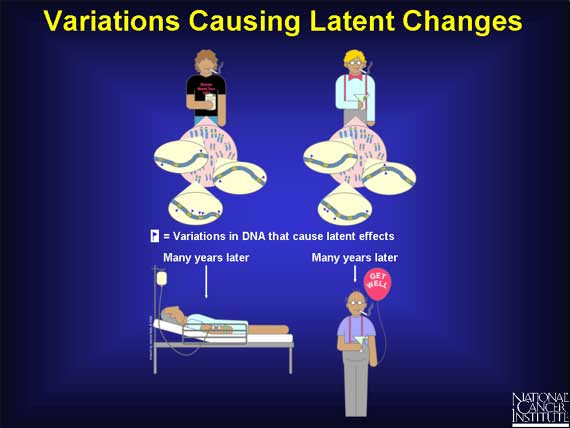
Finally, there are genetic variations that have "latent" effects. These variations, found in coding and regulatory regions, are not harmful on their own, and the change in each gene only becomes apparent under certain conditions. Such changes may eventually cause some people to be at higher risk for cancer, but only after exposure to certain environmental agents. They may also explain why one person responds to a drug treatment while another does not.
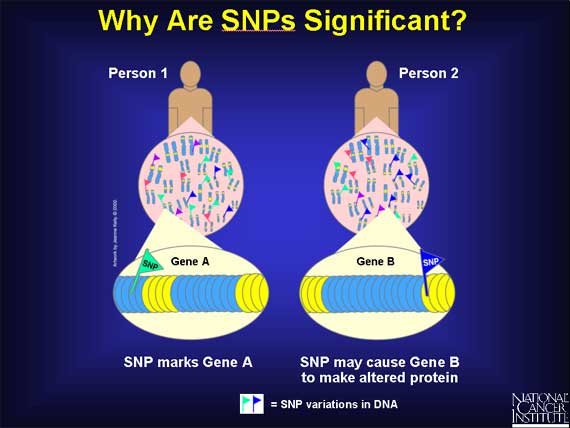
Here is part of the genome from two people who are both smokers and drinkers, but only one of them gets cancer. The zoom into the chromosomes of these two men shows just a sampling of the differences in variation that are responsible for their individual cancer risk. The variations themselves do not cause cancer. They only affect each person's susceptibility to tobacco smoke and alcohol after exposure.
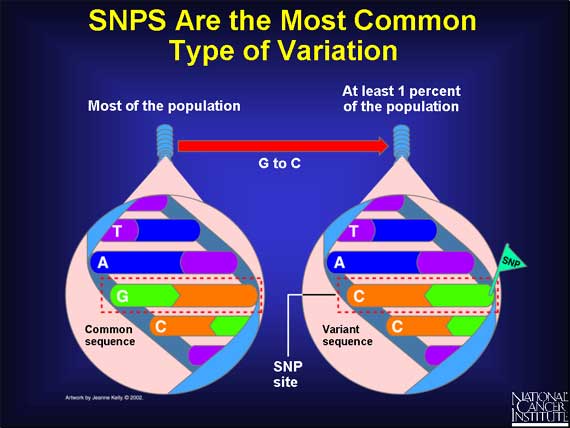
The most common type of genetic variation is called a Single Nucleotide Polymorphism (SNP).
A SNP is defined as a single base change in a DNA sequence that occurs in a significant proportion (more than 1 percent) of a large population. The single base is replaced by any of the other three bases. Here is an example: in the DNA sequence TAGC, a SNP occurs when the G base changes to a C, and the sequence becomes TACC.
SNPs are scattered throughout the genome and are found in both coding AND
noncoding regions. SNPs can cause silent, harmless, harmful, or latent effects.
They occur with a very high frequency, with estimates ranging from about 1 in
1000 bases to 1 in 100 to 300 bases. This means that there could be millions of
SNPs in each human genome. The abundance of SNPs and the ease with which they
can be measured make these genetic variations significant.
Most SNPs occur in noncoding regions and do not alter genes. Scientists are
finding that some of these SNPs have a useful function. If a SNP is frequently
found close to a particular gene, it acts as a marker for that gene.
The remaining SNPs occur in coding regions. They could alter the protein made
by that coding region, which in turn could influence a person's health. To
understand how a SNP could do this requires a brief look at proteins and their
building blocks, amino acids.
Humans use 20 different amino acids as building blocks to make the thousands of proteins found within the body. Each amino acid has the same basic structure with a central carbon atom that has four groups attached. Three of the groups are the same in every amino acid: one is a single hydrogen, one is an amino group that acts like a hook, and one is a carboxyl group that acts like an eye. The fourth group is a side chain, which is unique for every amino acid.
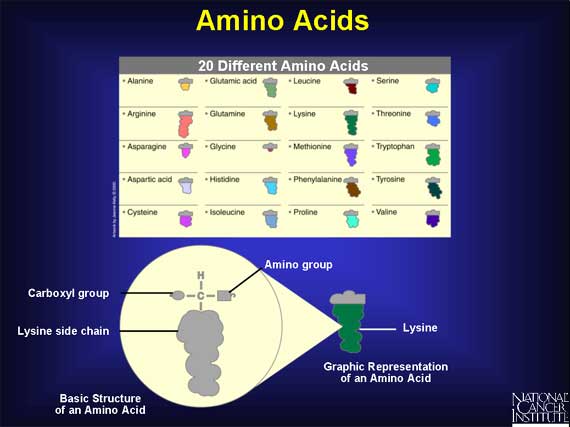
Humans use 20 different amino acids as building blocks to make the thousands of proteins found within the body. Each amino acid has the same basic structure with a central carbon atom that has four groups attached. Three of the groups are the same in every amino acid: one is a single hydrogen, one is an amino group that acts like a hook, and one is a carboxyl group that acts like an eye. The fourth group is a side chain, which is unique for every amino acid.
Each side chain has a unique size and shape: some side chains are round and bulky, some are short and skinny, and some are longer than others.
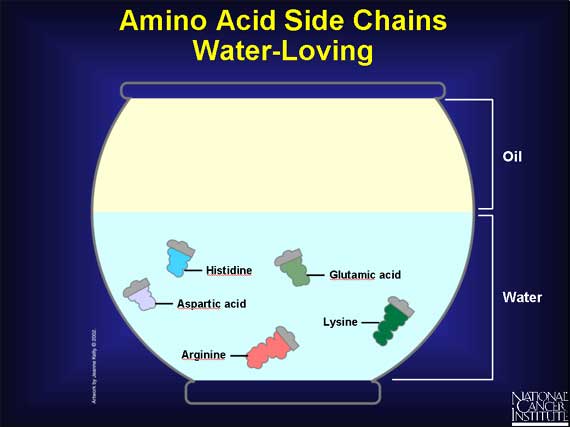
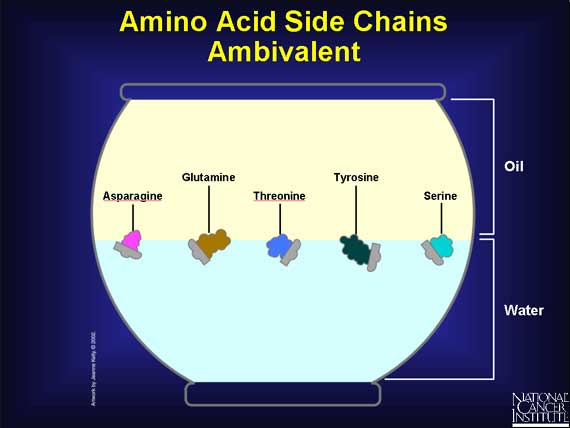
Unlike the carboxyl and amino groups, which always like a watery environment, side chains vary in their chemical properties.
--Some side chains have a charge, and that makes them prefer to be in water.
--Other side groups have no charge, and that makes them prefer an oily environment. It also prompts them to huddle close together.
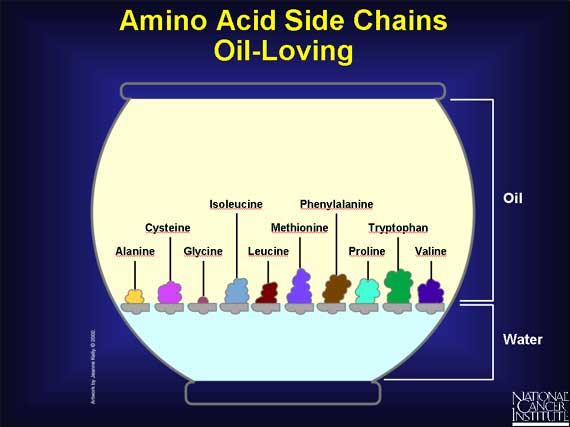
--And some side chains are downright indecisive. They usually prefer a watery environment, but tend to adjust themselves to oily ones as well.
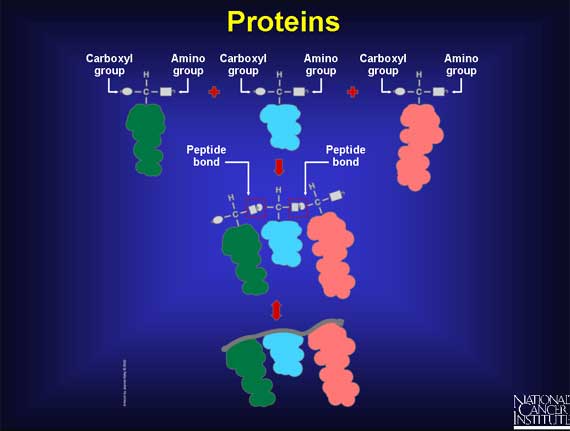
When two amino acids are lined up end-to-end, and the hooks and eyes are allowed to react together, a peptide bond forms. As more amino acids attach, a chain begins to form that has a strong but flexible backbone with side chains sticking out at regular intervals. This is protein-building in progress.
But how does the cell do this?
How does a sequence of DNA bases turn into a chain of amino acids and form a protein?
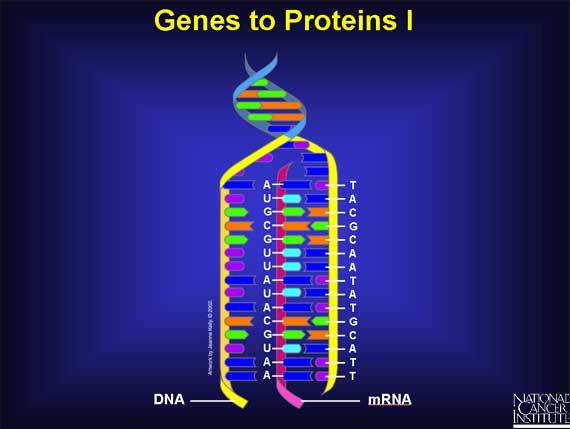
To turn a gene's DNA sequence of bases into a protein's sequence of amino acids requires an intermediary molecule called mRNA.
Base pairing is used to create mRNA from one of the DNA strands. Every T in the DNA strand pairs with an A in the mRNA strand, and Cs and Gs in the DNA pair with Gs and Cs in the mRNA. However, if there is an A in the DNA sequence, it pairs with a new base called Uracil (U) in the mRNA strand. Thus, if the DNA sequence is TACGCAATATGCATT, the mRNA sequence becomes AUGCGUUAUACGUAA.
The sequence of the bases of an mRNA (A, U, G, C) directly spells out the sequence of amino acids in the protein.
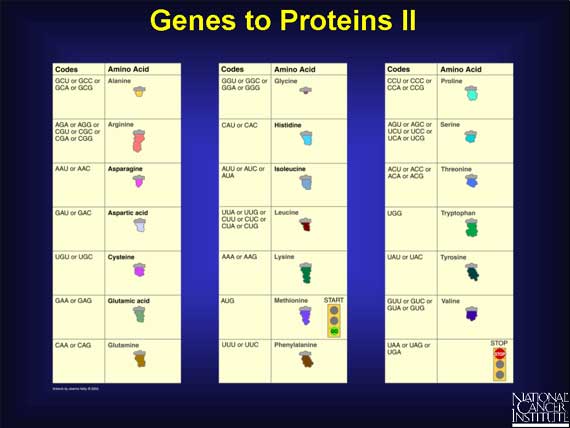
Every three bases in the mRNA sequence codes for a single amino acid and is called a "codon." There are 64 different ways to arrange the four bases in mRNA, e.g., UCA, UCG, AAA, and AGC, and yet there are only 20 different amino acids. This means that one amino acid can have more than one codon. For example, valine has four codons (GUU, GUC, GUA, and GUG), while histidine only has two (CAU and CAC).
The mRNA code also has one "start" and three "stop" signals. The codon AUG codes for an amino acid called methionine that always signals the start of protein building. Three other codons--UAG, UGA, and UAA--do not code for any amino acid. Instead, they always signal the mRNA to stop protein building.
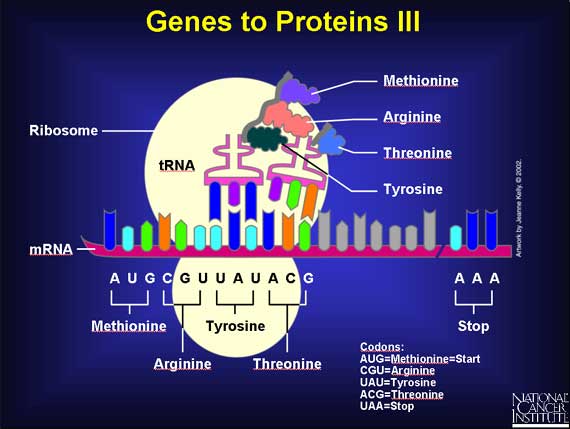
Proteins are built within a cell structure called a ribosome - a protein factory. First the mRNA attaches itself to the ribosome. Then specific adapter molecules, called transfer RNAs (tRNAs), match themselves to their appropriate codons. Each codon has a corresponding tRNA that carries a specific amino acid at one end.
Here is the beginning of a new protein. The first tRNA carrying methionine matched itself to the first codon. Then a second tRNA carrying arginine matched itself with the second codon. A chemical reaction joined one free end (the hook) of the methionine molecule to a free end (the eye) of the arginine molecule, and methionine's tRNA was released.
Now a third tRNA carrying tyrosine has matched the third codon, and a chemical reaction has joined the free end of the arginine to tyrosine, and arginine's tRNA has been released. The protein now has three amino acids joined together. This process will continue until a "stop" codon is reached.
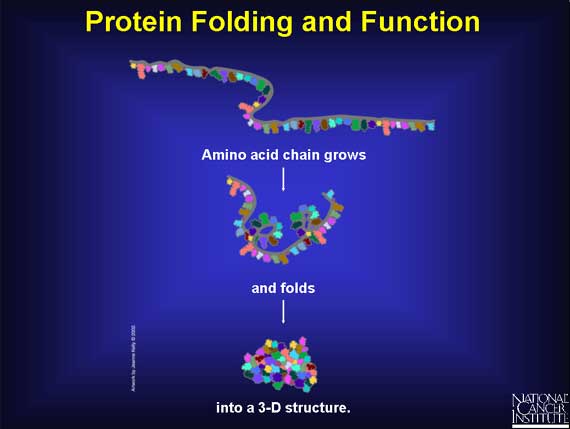
As the amino acid chain grows, it folds into a three-dimensional (3-D) structure, which depends on both the chemical nature and order of the different amino acids. The 3-D structure determines the function of the protein. When there is a change in one or more amino acids, then the ability of the protein to function may be affected. The protein's function may be unchanged or it may become sluggish, hyperactive, or inactive.
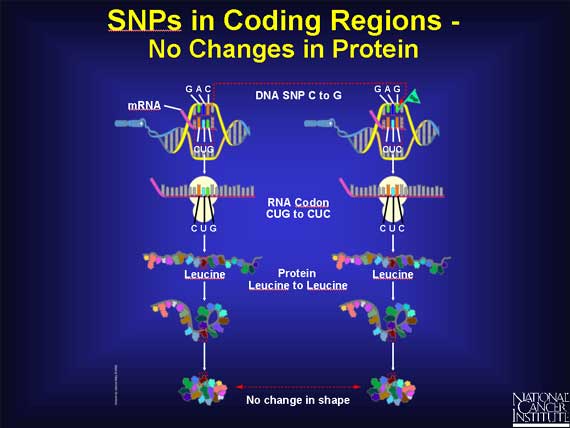
When a SNP occurs within a coding region, it is possible that there is no effect. For example, when the DNA sequence GAC becomes GAG, the codon in the mRNA changes from CUG to CUC. Since both codons stand for the same amino acid leucine, there is no change in the protein. The change in the DNA is called a silent change.
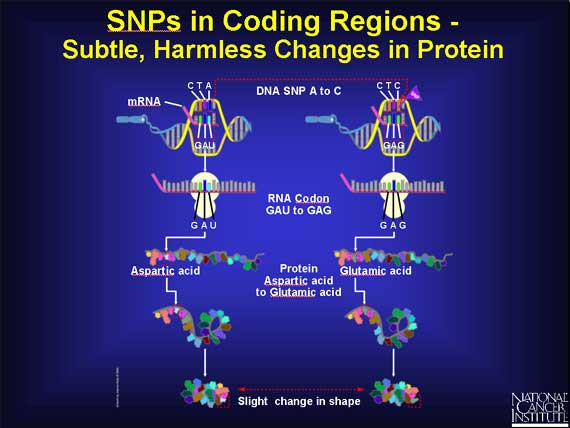
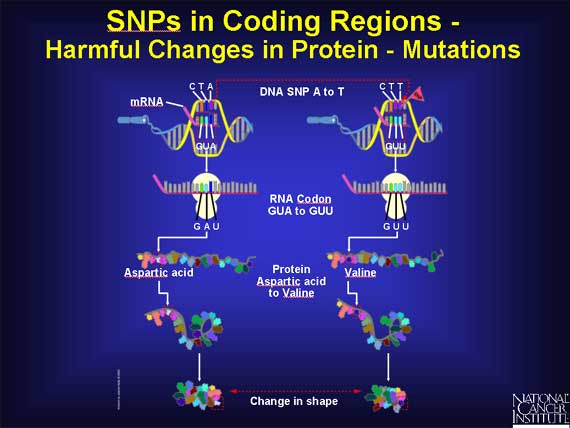
There are also SNPs within coding regions that lead to very subtle changes in proteins. For example, if a SNP causes the codon GAU to change to GAG, the amino acid changes from aspartic acid to glutamic acid. These amino acids have very similar chemical properties, but glutamic acid is just a little bigger. If the SNP causes a change in a part of the protein that is not important to its function, the result may be very subtle or totally harmless. The cell continues to function normally.
In another codon, the sequence GUA becomes GUU, and the amino acid called aspartic acid changes to valine in the protein. These two amino acids have very different chemical properties. The substitution of one for the other may, or may not, severely alter how the protein folds and functions, depending on where the change occurs in the protein. When the change in the protein does lead to disease symptoms in the patient, the SNP is harmful and is called a mutation.
An example of a SNP causing disease is found in sickle cell anemia. The change in one nucleotide base in the coding region for the hemoglobin beta gene causes the amino acid glutamic acid to be replaced by valine. As a result, the hemoglobin molecule can no longer carry oxygen as efficiently because the structure of the protein is changed dramatically.
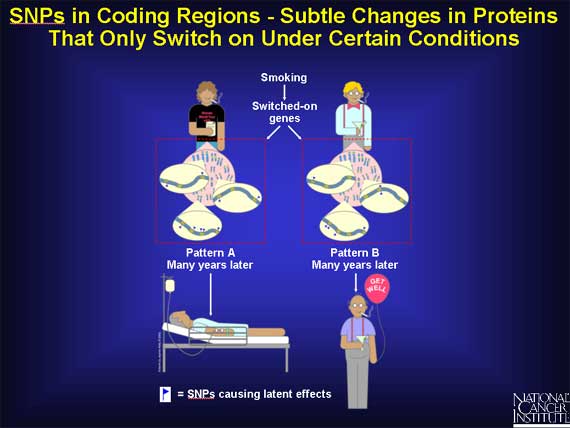
SNPs may cause subtle changes in a group of genes that under normal conditions are latent, i.e., they are switched "off." But when a person is exposed to precarcinogens or carcinogens, they can be switched "on."
Since the proteins from these genes regulate how fast or how slowly the harmful agents are absorbed, bound, metabolized, and excreted from the body, even a small or subtle change in any one of them may alter a person's risk for cancer.
*It is important to remember that the SNP itself does no harm under normal circumstances. Only when a person is exposed to an environmental agent that is carcinogenic, does the SNP exert an influence.
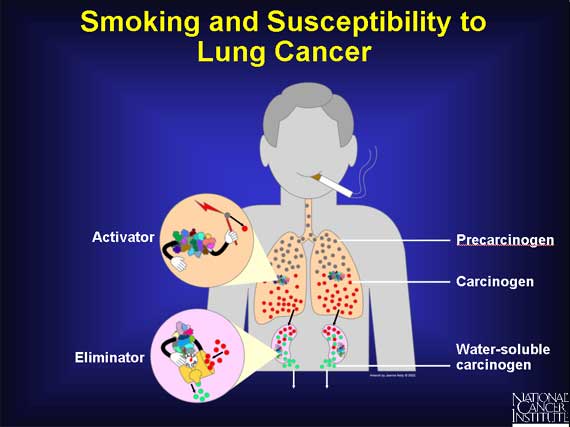
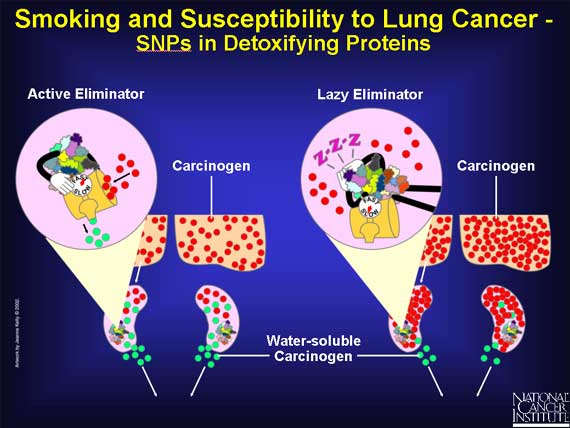
When a person smokes cigarettes, precarcinogens in the form of tobacco smoke enter a person's lungs and lodge in the fat-soluble area of the cells. They become bound to proteins that convert the precarcinogens into carcinogens. These reactive molecules are quickly handed over to detoxifying proteins that make the carcinogens water-soluble and allow the body to eliminate them into the urine, before they can damage the cell.
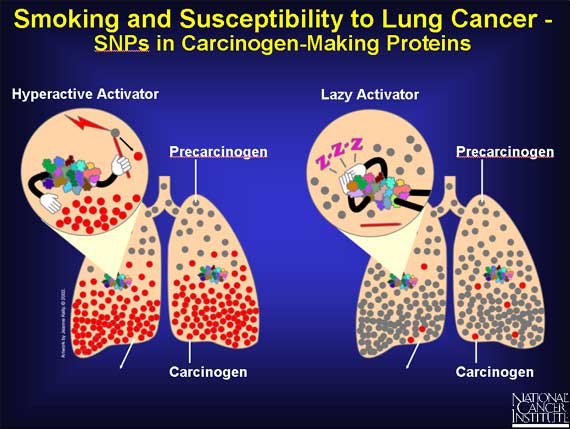
Because of SNPs, a person's genome may express a very active carcinogen-making protein, or a sluggish one, or something in between.
A protein with a very active binding site can "grab" more precarcinogen than usual. Or the protein may convert the precarcinogen to the carcinogen at a faster rate. In both cases, more carcinogen molecules pile up in the lungs, causing damage to the cells' DNA, which can lead to cancer.
On the other hand, if the SNPs result in a slow carcinogen-making protein, the lung is exposed to fewer DNA-damaging carcinogens, and the chance of cancer is reduced.
But until the body is exposed to cigarette smoke, no effect is seen.
SNPs may also influence detoxifying enzymes that prepare carcinogens for elimination from the human body. SNPs that result in very active forms of detoxifying enzymes will remove the carcinogens quickly from the body, allowing less time for damage.
SNPs that produce sluggish detoxifying enzymes will permit carcinogens to remain in the body for a longer time.
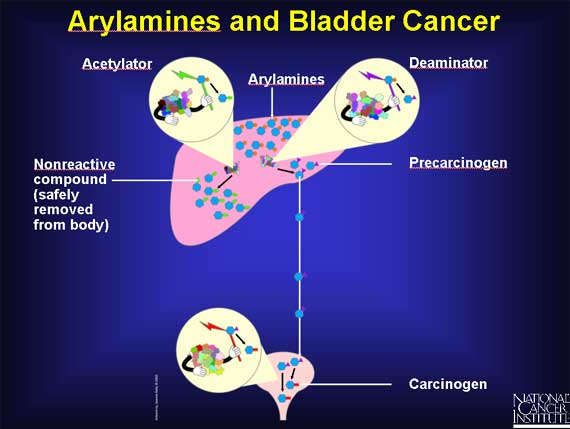
Some workers in the dye industry, after occupational exposure to arylamines, develop an increased risk of bladder cancer. Scientists suspect that SNPs may be involved.
In the liver, arylamines can have two fates, because there are two enzymes available to act on them.
An acetylator enzyme can deactivate arylamines, converting them into nonreactive compounds that are safely removed from the body.
Or arylamines can be activated by a deaminator enzyme in the liver to become precarcinogens that are carried to the bladder. There they are converted into carcinogens.
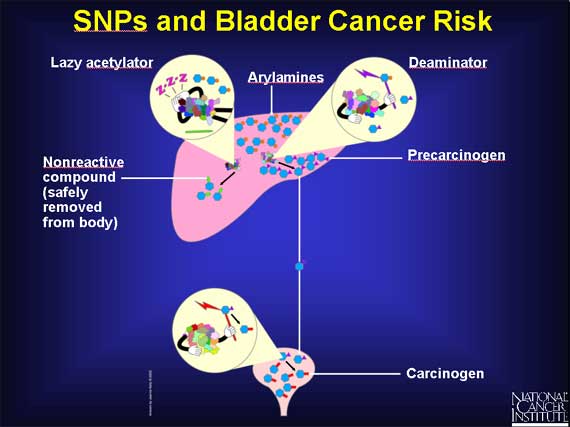
SNPs are responsible for the existence of several different slow forms of the acetylator enzyme. In individuals with a slow acetylator, the arylamines remain in the liver for a longer time, and more of them are converted to precarcinogens rather than nonreactive compounds. Thus, individuals with slow acetylators may be at increased risk for bladder cancer because they are exposed to more carcinogens in the bladder.
 |
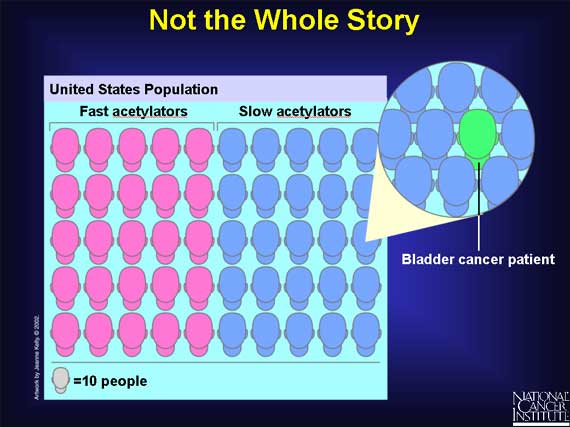
But SNPs are not always the entire story. Even among persons with slow acetylators who are exposed to arylamines, not everyone is at increased risk for bladder cancer. Research studies show that half the population in the United States have slow acetylators, and yet far fewer people, about one in 500, get bladder cancer. So having a "slow acetylator" does not automatically mean that you will get bladder cancer after arylamine exposure. Some other genes and their proteins must also be involved in affecting a person's risk for the disease. Researchers have yet to discover the full set of "risk" genes involved.
SNPs may also explain why some patients respond well to a specific drug treatment, while others have minimal or no response. SNPS may also be involved when patients have different side effects in response to the same drug.
Many proteins interact with the drug - involved in its transportation throughout the body, absorption into tissues, metabolism into more active forms or toxic by-products, and excretion. If a patient has SNPs in any one or more of these proteins, they may alter the time the body is exposed to active forms of the drug or any of its toxic byproducts.
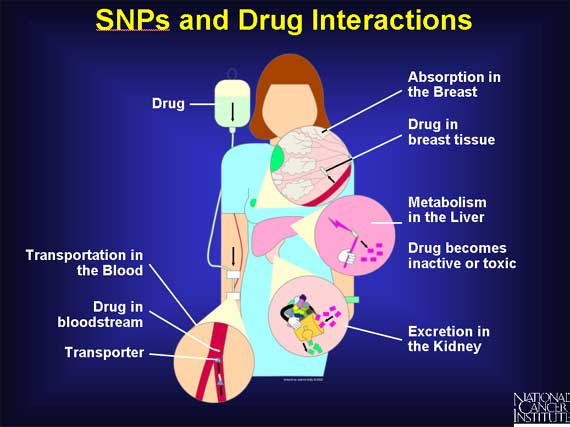
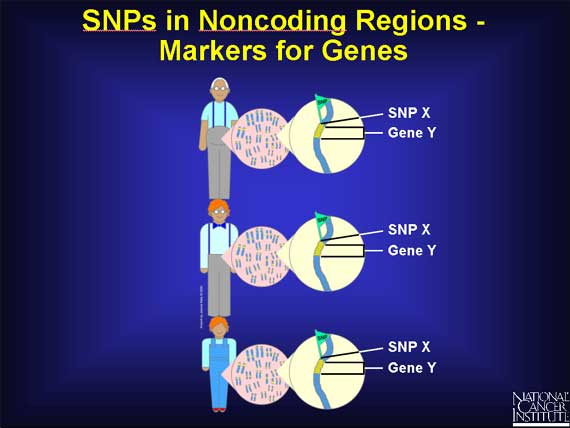
SNPs are very common variations scattered throughout the genome. Because they are fairly easy to measure and are also remarkably stable, being inherited from generation to generation, they have become useful as gene "markers." If a particular SNP is located near a gene, then every time that gene is passed from parent to child, the SNP is passed on also. This enables researchers to assume that when they find the same SNP in a group of individual genomes, the associated gene is present also.
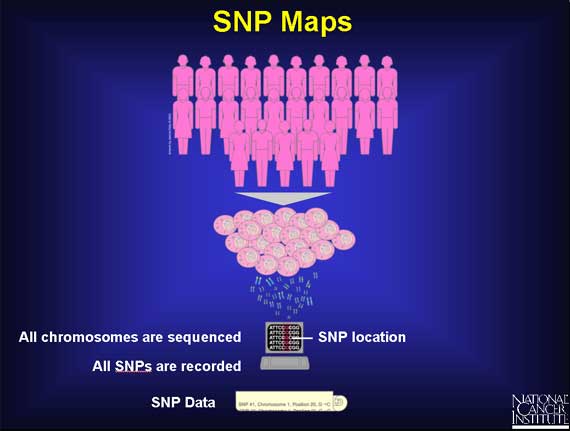
Scientists are trying to identify all the different SNPs in the human genome. They are sequencing the genomes of a large number of people and then comparing the base sequences to discover SNPs. The sequence data is being stored in computers that can generate a single map of the human genome containing all possible SNPs.
The genome of each individual contains its own pattern of SNPs. Thus, each individual has his or her own SNP profile. When scientists look at all the patterns from a large number of people they can organize them into groups.
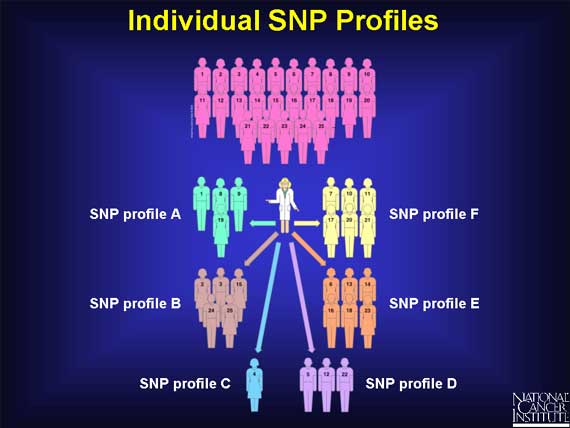
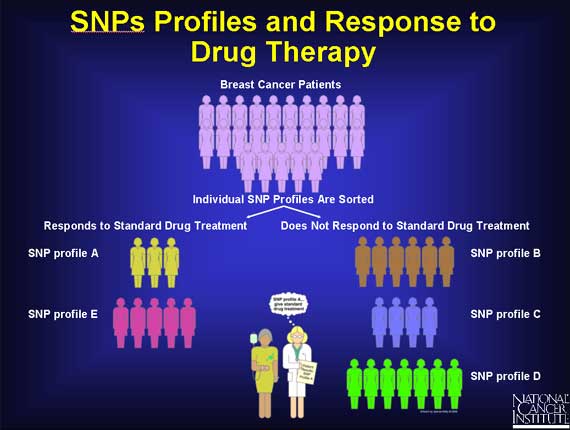
Doctors hope that by studying SNP profiles in populations, correlations will emerge between certain SNP profiles and specific responses to cancer treatment.
In the earlier example of two breast cancer patients, one did respond to the specific drug treatment, and the other did not. It may soon be possible to show that an entire group of patients responding well to breast cancer treatment repeatedly display profiles within a recurring set (e.g., A and E), while a group of non-responders repeatedly display profiles outside the set. In the future, after a doctor has diagnosed breast cancer in a new patient, he may also request a SNP profile and use the information to help advise his patient about her treatment options.
 |
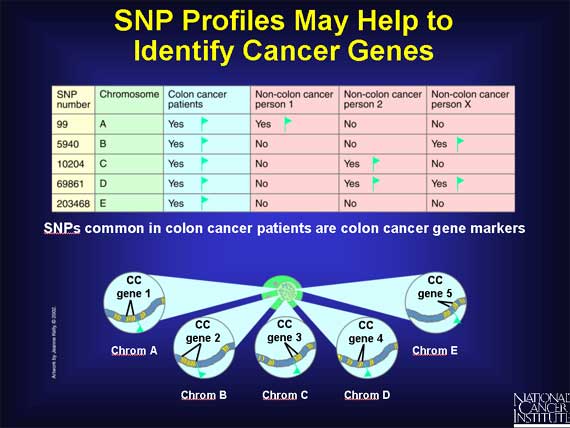
Since SNPs can be gene markers, SNP profiles may help scientists to identify the full collection of genes that contribute to the development of complex diseases such as cancer. For example, scientists will examine the SNP profiles from many people, including colon cancer patients. Within the many different profiles they may discover a small subset of SNPs that are only present in colon cancer patients. This implies that this subset of SNPs is associated only with colon cancer. When this happens, the SNPs themselves may be considered markers for colon cancer genes.
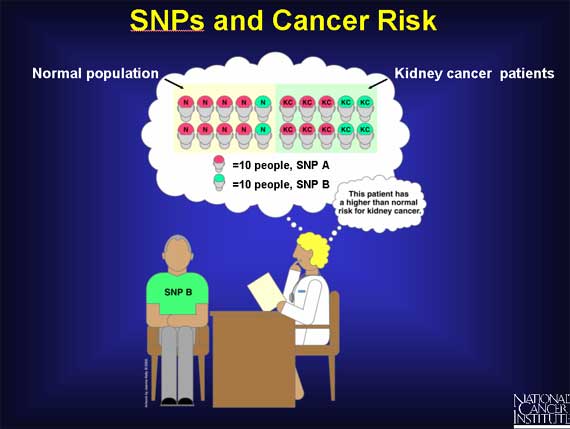
Scientists are also using SNPs to calculate risk factors associated with cancer in large populations.
First, imagine analyzing the SNPs in a random population of 100 people. 80 percent are found to have SNP A and the remaining 20 percent have SNP B. Now look at another 100 people, all with kidney cancer. In this group, 60 percent have SNP A and 40 percent have SNP B. Neither SNP A nor SNP B causes cancer. However, from this data, physicians can say that a person who has SNP B is at a higher risk for kidney cancer than a person with SNP A.
Armed with data from the SNP Map, cancer researchers across the country are looking for correlations between SNPs and precancerous conditions, SNPs and drug resistance in chemotherapy, SNPs and cancer susceptibility, and SNPs and drug response.
There is much work ahead, but there is also much hope that these and other research findings will result in improved health care for all.
|