ISSN: 1080-6059
Volume 14, Number 2–February 2008
Research
Molecular Typing of Australian Scedosporium Isolates Showing Genetic Variability and Numerous S. aurantiacum
Laurence Delhaes,*1,2 Azian Harun,*†1 Sharon
C.A. Chen,*1 Quoc Nguyen,‡1 Monica Slavin,§ Christopher
H. Heath,¶# Krystyna Maszewska,*† Catriona Halliday,* Vincent Robert,**
Tania C. Sorrell,* the Australian Scedosporium (AUSCEDO) Study Group,3 and
Wieland Meyer*†1 ![]()
*Westmead Hospital, Westmead, New South Wales, Australia; †University
of Sydney, Sydney, New South Wales, Australia; ‡St. Vincent's Hospital,
Sydney, New South Wales, Australia; §Alfred Hospital, Melbourne, Victoria,
Australia; ¶Royal Perth Hospital, Perth, Western Australia, Australia; #University
of Western Australia, Perth, Western Australia, Australia; and **Centraalbureau
voor Schimmelcultures, Utrecht, the Netherlands
Suggested citation for this article
Abstract
One hundred clinical isolates from a prospective nationwide study of scedosporiosis
in Australia (2003–2005) and 46 additional isolates were genotyped by
internal transcribed spacer–restriction fragment length polymorphism
(ITS-RFLP) analysis, ITS sequencing, and M13 PCR fingerprinting. ITS-RFLP and
PCR fingerprinting identified 3 distinct genetic groups. The first group corresponded
to Scedosporium prolificans (n = 83), and the other 2 comprised isolates
previously identified as S. apiospermum: one of these corresponded to S.
apiospermum (n = 33) and the other to the newly described species S.
aurantiacum (n = 30). Intraspecies variation was highest for S. apiospermum (58%),
followed by S. prolificans (45%) and S. aurantiacum (28%) as
determined by PCR fingerprinting. ITS sequence variation of 2.2% was
observed among S. apiospermum isolates. No correlation was found between
genotype of strains and their geographic origin, body site from which they
were cultured, or colonization versus invasive disease. Twelve S. prolificans isolates
from 2 suspected case clusters were examined by amplified fragment length polymorphism
analysis. No specific clusters were confirmed.
Despite efforts to identify and eliminate infectious agents, they continue to emerge and reemerge (1). Among them, pathogenic fungi contribute substantially to illness and death, especially in immunocompromised patients (2,3). In contrast to the well-documented opportunists Candida albicans, Cryptococcus neoformans, and Aspergillus fumigatus, the epidemiology and evolution of human infections caused by uncommon but emerging fungi are incompletely understood. Such pathogens include Scedosporium apiospermum (teleomorph Pseudallescheria boydii) and S. prolificans, which are inherently resistant to many antifungal agents (3–5).
S. apiospermum infections occur worldwide, ranging from localized mycetomas to deep-seated disease such as cerebral abscesses (6,7). This species also colonizes the respiratory tract of ≈10% of patients with cystic fibrosis and chronic suppurative lung disease (8–10). On the basis of genetic data, a new species, S. aurantiacum, was proposed for a subset of isolates previously identified as S. apiospermum (11). S. prolificans infections are geographically more restricted than those caused by S. apiospermum, being most prevalent in Australia, Spain, and the United States (12–15). S. prolificans typically causes localized infections in immunocompetent hosts but rapidly fatal disseminated infections in the immunocompromised among whom it has been associated with nosocomial outbreaks (3,12–17).
Since scedosporiosis, in particular that caused by S. prolificans, is often refractory to treatment (3,5), preventive strategies are of paramount importance. However, the epidemiology and mode of transmission of infection are not well understood. Furthermore, the environmental reservoir of S. prolificans is unknown. Molecular typing techniques now provide the means to elucidate the epidemiology of Scedosporium infections and to investigate potential case clusters (16,18,19). Strains recovered from patients with cystic fibrosis have demonstrated a high degree of genetic variability (10,20), although a single genetic profile predominated in 1 study (8). The degree of genetic variation within S. prolificans is more controversial. Two studies have reported low to no intraspecies genetic heterogeneity (16,21), while a third noted substantial genetic diversity (19). The results of these studies may be biased because they included only small numbers of isolates from specific patient populations. Genetic variability among S. aurantiacum has not yet been studied.
In this study, we used 4 molecular tools to examine genetic variation among a large number of Australian clinical Scedosporium isolates: 1) internal transcribed spacer (ITS)–based restriction fragment length polymorphism (ITS-RFLP) analysis; 2) DNA sequence analysis of the ITS region (selected isolates); 3) PCR fingerprinting using the microsatellite specific core sequence of phage M13; and 4) amplified fragment length polymorphism (AFLP) analysis (isolates from suspected case clusters). We also searched for the newly described species, S. aurantiacum and for genetic clustering of strains according to their geographic origin, body site from which they were cultured, and ability to cause invasive disease.
Materials and Methods
Scedosporium Isolates and Data Collection
A total of 146 Scedosporium isolates were studied (Technical
Appendix [![]() 43 KB, 6 pages]). Forty-six
were from the culture collection at the Clinical Mycology Laboratory, Centre
for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital,
Sydney, Australia. For these isolates, the following data were captured: demographic
information, patient coexisting conditions and risk factors (summarized in
the Technical Appendix [
43 KB, 6 pages]). Forty-six
were from the culture collection at the Clinical Mycology Laboratory, Centre
for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital,
Sydney, Australia. For these isolates, the following data were captured: demographic
information, patient coexisting conditions and risk factors (summarized in
the Technical Appendix [![]() 43 KB, 6 pages]). The remaining 100 isolates were obtained through
a national, prospective, laboratory-based surveillance for scedosporiosis in
Australia (the Australian Scedosporium [AUSCEDO] Study) from January
2003 to December 2005. The following data were collected: clinical status,
risk factor (defined according to published risk factors for scedosporiosis
[4,12–15]), major comorbidity (based on the International Classification
of Diseases, 10th revision, Australian Modification [ICD-10 AM] diagnostic
classification system [22]), isolated species, treatment and outcome. Scedosporium strains
obtained from a single colony from the primary isolation plate from all patients
were forwarded to the Molecular Mycology Research Laboratory, Westmead Hospital,
for genotyping. Isolates were identified as S. prolificans or S.
apiospermum by standard phenotypic methods (23). Species were confirmed
as S. prolificans or S. apiospermum, and S. aurantiacum was
identified (11) by ITS-RFLP analysis.
43 KB, 6 pages]). The remaining 100 isolates were obtained through
a national, prospective, laboratory-based surveillance for scedosporiosis in
Australia (the Australian Scedosporium [AUSCEDO] Study) from January
2003 to December 2005. The following data were collected: clinical status,
risk factor (defined according to published risk factors for scedosporiosis
[4,12–15]), major comorbidity (based on the International Classification
of Diseases, 10th revision, Australian Modification [ICD-10 AM] diagnostic
classification system [22]), isolated species, treatment and outcome. Scedosporium strains
obtained from a single colony from the primary isolation plate from all patients
were forwarded to the Molecular Mycology Research Laboratory, Westmead Hospital,
for genotyping. Isolates were identified as S. prolificans or S.
apiospermum by standard phenotypic methods (23). Species were confirmed
as S. prolificans or S. apiospermum, and S. aurantiacum was
identified (11) by ITS-RFLP analysis.
Definitions
An episode of scedosporiosis was defined as the incident isolation of Scedosporium spp. from any body site. Two or more episodes, fulfilling the case definition and occurring in different patients that were epidemiologically linked were defined as a potential case cluster. Invasive disease was defined according to the European Organization for Treatment of Cancer/Mycoses Study Group criteria for "definite" or "probable" infection (24). All other patients not fulfilling these criteria, including those with "possible" infection were considered colonized. Coincident hospital renovations or construction was considered to be a potential risk factor if major work was undertaken within 3 months before the isolation of Scedosporium spp. from a patient.
Description of 2 Potential Case Clusters
The first potential case cluster involved 8 patients located in the same hematology/hemopoietic
stem cell transplant (HSCT) unit at the Alfred Hospital, a large university
hospital in Melbourne (September 2000–October 2001; [15]). The
second consisted of 3 patients located in the same hematology/HSCT ward at
Westmead Hospital a major university hospital in Sydney (September 2003–January
2004; unpub. data). Details of the patients involved in these suspected case
clusters are summarized in the Technical Appendix [![]() 43 KB, 6 pages]). On each occasion,
patient isolates were submitted for genetic analyses to inform infection control
responses (see Results).
43 KB, 6 pages]). On each occasion,
patient isolates were submitted for genetic analyses to inform infection control
responses (see Results).
Genomic DNA Extraction and ITS-RFLP Analysis
Genomic DNA was isolated as described previously (18). The ITS1, 5.8S, and ITS2 regions of the rDNA gene cluster were amplified with the primers SR6R and LR1 (Table 1) as described previously (25). Amplicons were double digested with the restriction endonucleases Sau96I and HhaI (New England BioLabs, Ipswich, MA, USA) in accordance with the manufacturer's recommendations. Digested products were separated by electrophoresis in 3% agarose gels at 100 V for 3–4 h. Banding patterns were analyzed visually.
ITS Sequencing
Eleven isolates, representative of each of 3 ITS-RFLP patterns obtained, were selected for ITS sequencing: ITS-RFLP profile A (S. prolificans, WM 06.378, WM 06.440, and WM 06.393), ITS-RFLP profile B (S. apiospermum, WM 06.389, WM 06.471, and WM 06.497), and ITS-RFLP profile C (S. aurantiacum, WM 06.388, WM 06.482, WM 06.495, WM 06.496, and WM 06.498). The ITS region was amplified as described above and commercially sequenced in both directions by using SR6R or LR1 (Table 1) as forward and reverse primers.
PCR Fingerprinting
The minisatellite-specific core sequence of the wild-type phage M13 was used as a single primer for PCR fingerprinting (Table 1). Amplification reactions were performed as previously described (18). Blank control tubes containing all reagents except template DNA were included for each run; each sample was analyzed at least twice. PCR products were separated by electrophoresis on 1.4% agarose gels at 60 V for 14 cm. Strains were defined to be identical if their PCR fingerprinting profiles had a similarity of >97% ( = 1 band difference). Reproducibility of the PCR fingerprinting technique was accessed by re-amplifying 1 strain of each of the 3 Scedosporium spp. with all PCR amplifications carried out and re-running those on each gel.
AFLP Analysis
AFLP analysis was performed as described previously by using either EcoRI-GT 6-FAM-labeled and MseI-GT or EcoRI-TC 6-FAM-labeled and MseI-CA as selective primer pairs (QIAGEN, Valencia, CA, USA; Table 1) (26). All samples were analyzed by using the ABI Prism 3730 system (Applied Biosystems, Foster City, CA, USA). Data collation, fragment sizing, and pattern analyses were performed with GeneMapper software version 3.5 (Applied Biosystems). Only electrophoregram peaks above 1,000 fluorescent units were scored for the presence or absence of bands of the same size (range 50–500 bp) relative to the GeneScan 500 LIZ DNA size standard (Applied Biosystems). Only bands detected in duplicate AFLP experiments were included in the analysis.
Data Analysis
Clinical Data
Statistical analysis was performed by using SPSS version 10.0.07 (SPSS, Chicago, IL, USA) and EpiInfo version 6.0 (Centers for Disease Control and Prevention, Atlanta, GA, USA). Proportions were compared by using the χ2 or Fisher exact test. A p value <0.05 was statistically significant.
ITS Sequences
ITS sequences obtained from 11 isolates (see above) were aligned with the ITS sequences of the following reference strains obtained from GenBank: S. apiospermum CBS 101.22 (accession no. AJ888435), S. aurantiacum FMR 8630 (accession no. AJ888440), S. aurantiacum IHEM 15458 (accession no. AJ888441) and S. prolificans CBS 114.90 (accession no. AY882369) as well as 2 outgroup sequences: Pseudallescheria africana CBS 311.72 (accession no. AJ888425), and Petriella setifera CBS 164.74 (accession no. AY882352). Phylogenetic analyses were performed by using PAUP* version 4.06.10 (27).
PCR Fingerprinting Patterns and AFLP Fragments
PCR fingerprinting patterns were analyzed by using the 1D gel analysis module (BioGalaxy [BioAware, Hannut, Belgium]) in BioloMICS version 7.5.30 (BioAware). Images were normalized for lane to-lane differences in mobility by the alignment of patterns obtained on multiple loadings of the 1kb DNA size marker (GIBCO-BRL, Gaithersburg, MD, USA). The unweighted-pair group method by using arithmetic averages and the procedures of Nei and Li (28), both implemented in BioloMICS, were used to generate dendograms based on the coefficient of similarity (29) between the isolates. In addition, principal coordinate analysis (PcoA; BioloMICS) was conducted to give an overall representation of the observed strain variation. AFLP fragments were analyzed with BioloMICS.
Results
A total of 146 Scedosporium isolates from 120 episodes (119 patients)
were studied (Technical Appendix [![]() 43 KB, 6 pages]). Demographic
data were available for 108 (90%) episodes and coexisting conditions and risk
factor data for 115 (95.8%). Most episodes were reported from New South Wales
(64.2%), followed by Victoria (19.2%) and Western Australia (9.2%). The male:
female ratio was 1.3: 1. The major patient coexisting conditions and known
risk factors for scedosporiosis are summarized in the Technical
Appendix (
43 KB, 6 pages]). Demographic
data were available for 108 (90%) episodes and coexisting conditions and risk
factor data for 115 (95.8%). Most episodes were reported from New South Wales
(64.2%), followed by Victoria (19.2%) and Western Australia (9.2%). The male:
female ratio was 1.3: 1. The major patient coexisting conditions and known
risk factors for scedosporiosis are summarized in the Technical
Appendix (![]() 43 KB, 6 pages). Thirty-nine
patients (32.7%) had no underlying medical condition. Coincident building construction
was noted in 27 cases (22.5%). Scedosporium isolates were associated with invasive
disease in 46 (38.3%) instances; the remaining 74 (61.7%) were isolated from
patients who were colonized (Table 2).
43 KB, 6 pages). Thirty-nine
patients (32.7%) had no underlying medical condition. Coincident building construction
was noted in 27 cases (22.5%). Scedosporium isolates were associated with invasive
disease in 46 (38.3%) instances; the remaining 74 (61.7%) were isolated from
patients who were colonized (Table 2).
Molecular Typing of Scedosporium Isolates
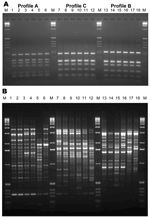 |
Figure 1. Internal transcribed spacer–restriction fragment length polymorphism (ITS-RFLP) patterns obtained by double digestion with the enzymes Sau96I and HhaI... |
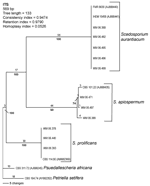 |
Figure 2. Rooted phylogram (outgroup Pseudallescheria africana CBS 311.72 and Petriella setifera CBS 164.74), showing the relationships among 11 selected strains... |
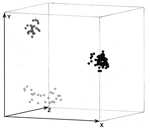 |
Figure 3. Three-dimensional presentation of the principal coordinate analysis of the PCR fingerprinting data showing 3 distinct clusters which correspond to Scedosporium prolificans (black dots)... |
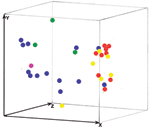 |
Figure 4. Three-dimensional presentation of the principal coordinate analysis of the combined M13 PCR fingerprinting, amplified fragment length polymorphism (AFLP) primers... |
 |
Appendix Figure 1. Dendogram generated from the PCR fingerprinting profiles obtained with the microsatellite primer M13 for all investigated Scedosporium isolates... |
 |
Appendix Figure 2. Dendogram generated by unweighted pair group method with arithmetic mean and the procedure of Nei and Li... |
All 146 isolates were examined by ITS-RFLP analysis and PCR fingerprinting. ITS sequencing was performed on 11 strains as described above. AFLP analysis was performed only for selected S. prolificans isolates, including the isolates of the suspected case clusters and isolates representative of the S. prolificans branches identified by PCR fingerprinting (Appendix Figure 1).
ITS-RFLP Analysis
RFLP analysis found 1 RFLP profile specific for S. prolificans isolates (ITS-RFLP profile A) and 2 profiles (ITS-RFLP profiles B and C) for isolates previously phenotypically identified as S. apiospermum (Figure 1, panel A). ITS-RFLP profile B corresponded to S. apiospermum and ITS-RFLP profile C to the newly described species, S. aurantiacum.
ITS Sequencing
Sequencing of the ITS 1, 5.8S, and ITS2 regions of the 11 strains, representative of each of the 3 ITS-RFLP profiles found the following results: BLAST searches against the corresponding GenBank reference sequences identified strains: WM 06.389 (accession no. EF639870), WM 06.497 (accession no. EF639872), and WM 06.471 (accession no. EF639871) (ITS-RFLP profile B) as S. apiospermum (96%–99% sequence similarity to strain CBS 101.22). Strains WM 06.388 (accession no. EF639865), WM 06.482 (accession no. EF639866), WM 06.495 (accession no. EF639867), WM 06.496 (accession no. EF639868), and WM 06.498 (accession no. EF639869) (ITS-RFLP profile C) were identified as S. aurantiacum (100% sequence identity with strains FMR 8630 and IHEM 15458). Isolates WM 06.393 (accession no. EF639863), WM 06.440 (accession no. EF639864) and WM 06.378 (accession no. EF639862) (ITS-RFLP profile A) were identified as S. prolificans (100% identity with strain CBS 114.90).
Phylogenetic analysis of the sequences demonstrated 3 distinct clades, the first corresponding to S. prolificans as the basal clade. The other 2 corresponded to the 2 more closely related but clearly distinct clades, S. apiospermum, and S. aurantiacum (Figure 2). S. apiospermum showed intraspecies sequence variation of 2.2% compared to S. aurantiacum and S. prolificans, which displayed no variation.
Final Identification of Scedosporium spp. and Clinical Associations
S. prolificans accounted for 75 patient episodes (83 of 146 isolates;
56.9%), S. apiospermum for 25 (33 isolates; 22.6%), and S.
aurantiacum for 23 (30 isolates; 20.6%) (Technical
Appendix [![]() 43 KB, 6 pages]). More
than 1 Scedosporium spp. was isolated from the same patient in 3 instances:
Patient 83: S. apiospermum (WM 06.471, WM 06.472, WM 06.474, and WM
06.475) and S. prolificans (WM 06.473); patient 91: S. apiospermum (WM
06.486) and S. prolificans (WM 06.485); and patient 102: S. apiospermum (WM
06.500) and S. prolificans (WM 06.501) (Technical
Appendix [
43 KB, 6 pages]). More
than 1 Scedosporium spp. was isolated from the same patient in 3 instances:
Patient 83: S. apiospermum (WM 06.471, WM 06.472, WM 06.474, and WM
06.475) and S. prolificans (WM 06.473); patient 91: S. apiospermum (WM
06.486) and S. prolificans (WM 06.485); and patient 102: S. apiospermum (WM
06.500) and S. prolificans (WM 06.501) (Technical
Appendix [![]() 43 KB, 6 pages]).
In 6 episodes, the same species was recovered from more than 1 body site in
the same patient at the same time (patients 57 [blood, bronchial washing, skin],
73 [blood, sputum], 80 [sputum, bone, wound fluid], 83 [bronchial washing,
bronchoalveolar lavage], 118 [pleural fluid, bone, wound fluid, chest tissue],
and 119 [blood, skin]; Technical Appendix [
43 KB, 6 pages]).
In 6 episodes, the same species was recovered from more than 1 body site in
the same patient at the same time (patients 57 [blood, bronchial washing, skin],
73 [blood, sputum], 80 [sputum, bone, wound fluid], 83 [bronchial washing,
bronchoalveolar lavage], 118 [pleural fluid, bone, wound fluid, chest tissue],
and 119 [blood, skin]; Technical Appendix [![]() 43 KB, 6 pages]).
43 KB, 6 pages]).
Approximately half (40%–52.2%) of S. apiospermum and S. aurantiacum isolates were from the respiratory tract/lung compared to 20% for S. prolificans. Conversely, all isolates from blood, 57.2% isolates from skin/soft tissue and 66.7% from eye were S. prolificans (Table 2). Invasive disease was more likely to be caused by S. prolificans than non-prolificans Scedosporium spp. (83% versus 17% of isolations; odds ratio (OR) 5.3, 95% confidence interval (CI) 2.0, 14.2, p = 0.002) (Table 2). This association was significant when compared with S. apiospermum as well as with S. aurantiacum (p<0.05; data not shown). The relative proportions of invasive disease among S. apiospermum and S. aurantiacum were similar (Table 2). Coincident building construction (27 cases, 22.5%) was more likely to be associated with isolation of S. prolificans compared with non-prolificans Scedosporium spp. (OR 11.5, 95% CI 2.4, 74.5; p<0.001; data not shown).
Molecular Epidemiology
Strain Typing
PCR fingerprinting delineated 3 major clusters concordant with S. apiospermum, S. aurantiacum, and S. prolificans (Appendix Figure 1; Figure 1, panel B; Figure 3). Clusters corresponding to S. aurantiacum and S. prolificans were substantially more densely grouped than the S. apiospermum cluster (Figure 3).
PCR fingerprinting profiles showed polymorphisms within each of the 3 species,
allowing for a clear differentiation, by using a "cut-off point" of >97%
similarity. Multiple isolates from the same patient obtained from different
anatomic sites (Technical Appendix [![]() 43 KB, 6 pages]) had identical
or >97%
similarity between their PCR fingerprints, except for 1 patient (patient 118).
In 8 instances, PCR fingerprinting showed that patients were infected with
2 different strains: (patients 1, 10, 27, 57, 83 99, 118 [Appendix
Figure 1, Technical Appendix (
43 KB, 6 pages]) had identical
or >97%
similarity between their PCR fingerprints, except for 1 patient (patient 118).
In 8 instances, PCR fingerprinting showed that patients were infected with
2 different strains: (patients 1, 10, 27, 57, 83 99, 118 [Appendix
Figure 1, Technical Appendix (![]() 43 KB, 6 pages)]). For all species,
genetic profiles were independent of geographic origin, body site of isolation
or whether the patient was infected or colonized (Appendix
Figure 1). Profiles
were also independent of patient comorbidityity and risk factors for scedosporiosis
(data not shown). Intraspecies PCR fingerprinting variation was highest for S.
apiospermum (58%)
followed by S. prolificans (45%) and S. aurantiacum (28%)
(Appendix Figure 1).
43 KB, 6 pages)]). For all species,
genetic profiles were independent of geographic origin, body site of isolation
or whether the patient was infected or colonized (Appendix
Figure 1). Profiles
were also independent of patient comorbidityity and risk factors for scedosporiosis
(data not shown). Intraspecies PCR fingerprinting variation was highest for S.
apiospermum (58%)
followed by S. prolificans (45%) and S. aurantiacum (28%)
(Appendix Figure 1).
Examination of Isolates from Suspected Case Clusters
Twelve isolates from 2 presumptive case clusters of S. prolificans infection
(Alfred Hospital, Melbourne patients: isolates WM 06.392, WM 06.393, WM 06.395,
WM 06.399, WM 06.400, WM 06.401, WM 06.402, and WM 06.405; Westmead Hospital,
Sydney patients: isolates WM 06.432, WM 06.434, WM06.457, and WM 06.458; Technical Appendix [![]() 43 KB, 6 pages]) as well as 23 additional isolates, representative of the S.
prolificans branches identified by PCR fingerprinting (Appendix
Figure 1) were further investigated by AFLP typing. S. prolificans was
not isolated from the environment in either setting despite extensive sampling.
The AFLP bands were found to be 50–493 bp by using the primers EcoRI-GT
and MseI-GT (data not shown), and from 52–468 bp by using the
primers EcoRI-TG and MseI-CA (Appendix
Figure 2). These 35 isolates exhibited
32 different AFLP profiles, with isolates from the same patient (patients 1,
73, and 119) showing identical profiles (Appendix Figure
2), confirming
the PCR fingerprinting results (Appendix Figure 1). PcoA of the combined
AFLP and PCR fingerprinting data demonstrated no clustering of these isolates
(Figure 4), which ruled out the possibility of nosocomial transmission.
43 KB, 6 pages]) as well as 23 additional isolates, representative of the S.
prolificans branches identified by PCR fingerprinting (Appendix
Figure 1) were further investigated by AFLP typing. S. prolificans was
not isolated from the environment in either setting despite extensive sampling.
The AFLP bands were found to be 50–493 bp by using the primers EcoRI-GT
and MseI-GT (data not shown), and from 52–468 bp by using the
primers EcoRI-TG and MseI-CA (Appendix
Figure 2). These 35 isolates exhibited
32 different AFLP profiles, with isolates from the same patient (patients 1,
73, and 119) showing identical profiles (Appendix Figure
2), confirming
the PCR fingerprinting results (Appendix Figure 1). PcoA of the combined
AFLP and PCR fingerprinting data demonstrated no clustering of these isolates
(Figure 4), which ruled out the possibility of nosocomial transmission.
Discussion
We examined genetic variation among a large number of population-derived Scedosporium isolates across the Australian continent. In line with previously reported genetic variability in the S. apiospermum/P. boydii species complex (30–32), we observed 2 distinct ITS-RFLP patterns among S. apiospermum isolates, showing the presence of the newly described species S. aurantiacum (11). Notably, we have identified by ITS sequencing that S. aurantiacum comprised 45% of the current collection of Australian "S. apiospermum" isolates and documents genetic variability within S. aurantiacum.
Epidemiologic investigation of Scedosporium infection requires accurate identification and typing. S. apiospermum, S. aurantiacum, and S. prolificans were clearly distinguished from each other by PCR fingerprinting and ITS-RFLP analysis. This is consistent with previous rDNA sequence-based studies (30,33,34). The observation of 2 distinct genetic groups, corresponding to S. aurantiacum and S. apiospermum, supports the proposal that S. aurantiacum be designated a separate species (11). This proposal is also supported by the 5%–10% ITS sequence variation found between S. aurantiacum and S. apiospermum compared to an absence of intraspecies variation in S. aurantiacum and S. prolificans and a 2.2% variation in S. apiospermum (30,32; current study).
Using PCR fingerprinting, intraspecies variation was greatest (58%) among S. apiospermum isolates (Figure 3). This diversity is generally consistent with the high degree of polymorphism (15–20 genotypes) previously found (10,20,32). In contrast, genetic variation was lowest (28%) among the S. aurantiacum isolates (Appendix Figure 1; Figure 3). Nevertheless, PCR fingerprinting polymorphisms clearly differentiated all 30 strains (Appendix Figure 1). Further genotyping studies of a greater number of and more geographically diverse S. aurantiacum isolates are warranted.
The intraspecies PCR fingerprint variation in S. prolificans (45%) was greater than that in S. aurantiacum but less than that in S. apiospermum. Given that S. aurantiacum is phylogenetically more closely related to S. apiospermum than to S. prolificans (11,33; current study), this result was unexpected. It may be due to different evolutionary pressures acting on the 3 different species or the relatively small numbers of S. aurantiacum isolates studied to date. The moderate genetic diversity among S. prolificans confirms previous findings (19). Despite the observed polymorphisms, PcoA of PCR fingerprint profiles showed dense clustering for S. prolificans (Figure 3), which is consistent with the low to absent intraspecies variability in S. prolificans found by others (20,21,33). These apparently contradictory findings emphasize the importance of choosing the optimum molecular typing tool with the most appropriate discriminatory power for the organism or species being studied.
The high degree of intraspecies variation detected by PCR fingerprinting and
AFLP analysis supports the use of these methods to establish genetic relatedness
between isolates recovered from different patients or multiple isolates from
the same patient. In comparison, the variation detected by ITS-RFLP analysis
and ITS sequencing corresponded to interspecies variation, which makes those
techniques ideal for identification of any given isolate to the species level.
Individual patients are most likely infected or colonized with genetically
distinct strains (19–21; this study). Identical PCR fingerprint
or AFLP profiles were noted in multiple isolates recovered simultaneously from
different anatomic sites in the same patient (21; current study).
However, 8 patients were infected or colonized by at least 2 strains as reflected
by their different genetic profiles (Technical
Appendix [![]() 43 KB, 6 pages]). Possible explanations
include concomitant infection by multiple strains from which only a restricted
number were recovered, or colonization by 1 strain followed by infection or
colonization with a second strain of a different genotype. Longitudinal genotyping
studies are required to determine the likelihood that persistence of >1
genotypes later leads to clinically important infection or whether the disease
is more likely to be caused by an unrelated genotype. In this context, the
development of a multilocus sequence typing scheme for Scedosporium,
as has been developed for Candida spp. (35), would be of great
advantage to overcome interlaboratory reproducibility problems, which are known
to be associated with PCR fingerprinting or AFLP data. However, developing
such a scheme remains cumbersome due to the current lack of genomic data of Scedosporium spp.
43 KB, 6 pages]). Possible explanations
include concomitant infection by multiple strains from which only a restricted
number were recovered, or colonization by 1 strain followed by infection or
colonization with a second strain of a different genotype. Longitudinal genotyping
studies are required to determine the likelihood that persistence of >1
genotypes later leads to clinically important infection or whether the disease
is more likely to be caused by an unrelated genotype. In this context, the
development of a multilocus sequence typing scheme for Scedosporium,
as has been developed for Candida spp. (35), would be of great
advantage to overcome interlaboratory reproducibility problems, which are known
to be associated with PCR fingerprinting or AFLP data. However, developing
such a scheme remains cumbersome due to the current lack of genomic data of Scedosporium spp.
For all 3 Scedosporium spp., there was no clustering of strains according to their geographic or body site of origin or by their ability to cause invasive disease, which is in agreement with previous findings for S. apiospermum (20,30) and S. prolificans (16,17,21). Of note, no specific genotypes were associated with underlying medical conditions or risk factors. Compared with S. apiospermum and S. aurantiacum S. prolificans was more frequently associated with coincident hospital renovation, and invasive disease, had a greater predilection to cause disseminated infection and was the predominant species isolated from blood and other sterile sites (12–16,36; current study). Our preliminary observations indicate that the epidemiology and clinical relevance of recovering S. aurantiacum may be similar to that of S. apiospermum. S. aurantiacum has been reported to colonize the respiratory tract of at-risk patients (8).
In addition to PCR fingerprinting, we applied AFLP analysis to investigate the possibility of 2 case clusters caused by S. prolificans. AFLP analysis was chosen as an independent technique using 2 combinations of selective primers (Table 1), which have been previously shown to have good discriminatory power for fungal strain differentiation (26). Both techniques, previously used to identify outbreak strain clusters in the recent cryptococcosis outbreak on Vancouver Island (37), generated in the current situation distinct patterns from all S. prolificans isolates except serial isolates obtained from the same patient (Appendix Figures 1, 2). These findings exclude the occurrence of nosocomial outbreaks or any close relationship with the nonoutbreak isolates, a result similar to those obtained previously (38). Overall nosocomial acquisition of infection has been demonstrated in only 2 instances (16,17). Scedosporium spp. have rarely been isolated from hospital air or from indoor or outdoor surface samples (13,39,40, current study), which raises questions about the mode of acquisition by patients and the mechanisms of the selection of this specific fungus as an infectious agent from among the high biodiversity of environmental molds.
In conclusion, ITS-RFLP analysis is a powerful tool for distinguishing between isolates of the new species S. aurantiacum and S. apiospermum. PCR fingerprinting and AFLP analysis are useful techniques for determining genetic relatedness between Scedosporium isolates and for investigating potential case clusters.
Acknowledgments
We thank all participating infectious disease physicians, clinical microbiologists, and hospital scientists. We also thank Nathalie van de Wiele for assistance in figure preparation, and Dee Carter and Tien Bui for sharing their AFLP expertise.
This study was supported by an NH&MRC project grant no. 352303 to W.M. and V.R., a Center for Infectious Diseases and Microbiology–Public Health start-up grant to W.M. and S.C.A.C., and a Merck, Sharp and Dohme, Australia grant to S.C.A.C. and W.M. This is a publication of the AUSCEDO and ECMM-ISHAM working groups on Pseudallescheria/Scedosporium infections.
S.C.A.C. is a member of the Antifungal Advisory Board of Gilead Sciences and Pfizer Australia. C.H.H., M.S., and T.C.S. are or have been on Antifungal Advisory Boards for Gilead Sciences; Pfizer Australia; Merck, Sharp and Dohme, Australia; and Schering-Plough, Australia. T.S. has been on a Global Advisory Board for Pfizer US. T.C.S., M.S., and S.C.A.C. have received untied project funding from Pfizer US; Pfizer Australia; Merck, Sharp and Dohme Australia; and Gilead Sciences. T.C.S. has also received funding from Merck US.
Dr Delhaes is the head of the filamentous fungal research group EA3609 at the Pasteur Institute in Lille, France. Her major research interests are systemic mycoses due to Aspergillus and Scedosporium, including molecular epidemiology, nosocomial aspects of the diseases, and oxidative stress response.
References
- Rappuoli R. From Pasteur to genomics: progress and challenges in infectious diseases. Nat Med. 2004;10:1177–85.
- Nucci M. Emerging moulds: Fusarium, Scedosporium and Zygomycetes in transplant patients. Curr Opin Infect Dis. 2003;16:607–12.
- Walsh TJ, Groll A, Hiemenez J, Fleming R, Roilides E, Anaissie E. Infections due to emerging and uncommon medically-important fungal pathogens. Clin Microbiol Infect. 2004;10(Suppl 1):48–66.
- Steinbach WJ, Perfect JR. Scedosporium species infections and treatments. J Chemother. 2003;15(Suppl 2):16–27.
- Gilgado F, Serena C, Cano J, Gene J, Guarro J. Antifungal susceptibilities of the species of the Pseudallescheria boydii complex. Antimicrob Agents Chemother. 2006;50:4211–3.
- Guarro J, Kantarcioglu AS, Horre R, Rodriguez-Tudela JL, Cuenca Estrella M, Berenguer J, et al. Scedosporium apiospermum: changing clinical spectrum of a therapy-refractory opportunist. Med Mycol. 2006;44:295–327.
- Campagnaro EL, Woodside KJ, Early MG, Gugliuzza KK, Colome-Grimmer MI, Lopez FA, et al. Disseminated Pseudallescheria boydii (Scedosporium apiospermum) infection in a renal transplant patient. Transpl Infect Dis. 2002;4:207–11.
- Williamson EC, Speers D, Arthur IH, Harnett G, Ryan G, Inglis TJ. Molecular epidemiology of Scedosporium apiospermum infection determined by PCR amplification of ribosomal intergenic spacer sequences in patients with chronic lung disease. J Clin Microbiol. 2001;39:47–50.
- Cimon B, Carrere J, Vinatier JF, Chazalette JP, Chabasse D, Bouchara JP. Clinical significance of Scedosporium apiospermum in patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis. 2000;19:53–6.
- Defontaine A, Zouhair R, Cimon B, Carrere J, Bailly E, Symoens F, et al. Genotyping study of Scedosporium apiospermum isolates from patients with cystic fibrosis. J Clin Microbiol. 2002;40:2108–14.
- Gilgado F, Cano J, Gene J, Guarro J. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J Clin Microbiol. 2005;43:4930–42.
- Wood GM, McCormack JG, Muir DB, Ellis DH, Ridley MF, Pritchard R, et al. Clinical features of human infection with Scedosporium inflatum. Clin Infect Dis. 1992;14:1027–33.
- Idigoras P, Perez-Trallero E, Pineiro L, Larruskain J, Lopez-Lopategui MC, Rodriguez N, et al. Disseminated infection and colonization by Scedosporium prolificans: a review of 18 cases, 1990–1999. Clin Infect Dis. 2001;32:e158–65.
- Berenguer J, Rodriguez-Tudela JL, Richard C, Alvarez M, Sanz M, Gaztelurrutia L, et al. Deep infections caused by Scedosporium prolificans. A report on 16 cases in Spain and a review of the literature. Scedosporium prolificans Spanish Study Group. Medicine (Baltimore). 1997;76:256–65.
- Cooley L, Spelman D, Thursky K, Slavin M. Infection with Scedosporium apiospermum and S. prolificans, Australia. Emerg Infect Dis. 2007;13:1170–7.
- Ruiz-Diez B, Martin-Diez F, Rodriguez-Tudela JL, Alvarez M, Martinez-Suarez JV. Use of random amplification of polymorphic DNA (RAPD) and PCR-fingerprinting for genotyping a Scedosporium prolificans (inflatum) outbreak in four leukemic patients. Curr Microbiol. 1997;35:186–90.
- Guerrero A, Torres P, Duran MT, Ruiz-Diez B, Rosales M, Rodriguez-Tudela JL. Airborne outbreak of nosocomial Scedosporium prolificans infection. Lancet. 2001;357:1267–8.
- Meyer W, Maszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by PCR-fingerprinting and RAPD—a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–9.
- Sole M, Cano J, Rodriguez-Tudela JL, Ponton J, Sutton DA, Perrie R, et al. Molecular typing of clinical and environmental isolates of Scedosporium prolificans by inter-simple-sequence-repeat polymerase chain reaction. Med Mycol. 2003;41:293–300.
- Zouhair R, Defontaine A, Ollivier C, Cimon B, Symoens F, Hallet J-N, et al. Typing of Scedosporium apiospermum by multilocus enzyme electrophoresis and random amplification of polymorphic DNA. J Med Microbiol. 2001;50:925–32.
- San Millan R, Quindos G, Garaizar J, Salesa R, Guarro J, Ponton J. Characterization of Scedosporium prolificans clinical isolates by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:2270–4.
- International statistical classification of disease and related health problems, 10th revision, Australian Modification (ICD-10-AM). Sydney (Australia): National Centre for Classification in Health, University of Sydney; 1998.
- de Hoog GS, Guarro J, Gene J, Figuerras MJ. Hyphomycetes: Genus: Scedosporium. In: Atlas of clinical fungi. 2nd ed. Utrecht (the Netherlands): Centralbureau voor Schimmelcultures/Universitat Rovira i Virgili; 2000. p. 899–901.
- Ascioglu S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, et al. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoeitic stem cell transplants: an international consensus. Clin Infect Dis. 2002;34:7–14.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–46.
- Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J Clin Microbiol. 2003;41:703–11.
- Swofford DL. PAUP* 4.06.10: Phylogenetic analysis using parsimony. Sunderland (MA): Sinauer Associates; 2003.
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76:5269–73.
- Wetton JH, Carter RE, Parkin DT, Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987;327:147–9.
- Rainer J, de Hoog GS, Wedde M, Graser Y, Gilges S. Molecular variability of Pseudallescheria boydii, a neurotropic opportunist. J Clin Microbiol. 2000;38:3267–73.
- de Hoog GS, Marvin-Sikkema FD, Lahpoor GA, Gottschall JC, Prins RA, Gueho E. Ecology and physiology of Pseudallescheria boydii, an emerging opportunistic fungus. Mycoses. 1994;37:71–8.
- Gueho E, de Hoog GS. Taxonomy of the medical species of Pseudallescheria and Scedosporium. J Mycol Méd. 1991;1:3–9.
- Rainer J, de Hoog GS. Molecular taxonomy and ecology of Pseudallescheria, Petriella and Scedosporium prolificans (Microascaceae) containing opportunistic agents on humans. Mycol Res. 2006;110:151–60.
- Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, Davidson AD, et al. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol. 2003;41:5265–6.
- Wedde M, Muller D, Tintelnot K, de Hoog GS, Stahl U. PCR-based identification of clinically relevant Pseudallescheria/Scedosporium strains. Med Mycol. 1998;36:61–7.
- Alvarez M, Lopez Ponga B, Rayon C, Garcia Gala J, Roson Porto MC, Gonzalez M, et al. Nosocomial outbreak caused by Scedosporium prolificans (inflatum): four fatal cases in leukemic patients. J Clin Microbiol. 1995;33:3290–5.
- Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101:17258–63.
- Tapia M, Richard C, Baro J, Salesa R, Figols J, Zurbano F, et al. Scedosporium inflatum infection in immunocompromised hematological patients. Br J Haematol. 1994;87:212–4.
- Idigoras P, Garcia-Arenzana JM, Saenz JR, Pineiro L, Marin J. Isolation of Scedosporium prolificans from the air in the room of a patient with leukemia and disseminated infection with this fungus. Enferm Infecc Microbiol Clin. 2000;18:426–7.
- Salkin IF, McGinnis MR, Dykstra MJ, Rinaldi MG. Scedosporium inflatum, an emerging pathogen. J Clin Microbiol. 1988;26:498–503.
Figures
Figure 1. Internal transcribed spacer–restriction fragment length polymorphism
(ITS-RFLP) patterns obtained by double digestion with the enzymes Sau96I
and HhaI...
Figure 2. Rooted phylogram (outgroup Pseudallescheria africana CBS
311.72 and Petriella setifera CBS 164.74), showing the relationships
among 11 selected strains...
Figure 3. Three-dimensional presentation of the principal coordinate analysis
of the PCR fingerprinting data showing 3 distinct clusters which correspond
to Scedosporium prolificans (black dots)...
Figure 4. Three-dimensional presentation of the principal coordinate analysis
of the combined M13 PCR fingerprinting, amplified fragment length polymorphism
(AFLP) primers...
Appendix Figure 1. Dendogram
generated from the PCR fingerprinting profiles obtained with the microsatellite
primer M13 for all investigated Scedosporium isolates...
Appendix Figure 2. Dendogram
generated by unweighted pair group method with arithmetic mean and the procedure
of Nei and Li...
Tables
Table 1. Primer and adaptor oligonucleotide sequences used in the study
Table 2. Selected characteristics for 120 isolations
(episodes) of Scedosporium spp.
Suggested Citation for this Article
Delhaes L, Harun A, Chen SCA, Nguyen Q, Slavin M, Heath CH, et al. Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum. Emerg Infect Dis [serial on the Internet]. 2008 Feb [date cited]. Available from http://www.cdc.gov/EID/content/14/2/282.htm
1These authors contributed equally to experimental work and data analysis.
2Current affiilation: Lille Pasteur Institute, Lille, France
3Members of the Australian Scedosporium Study Group of the Australasian Society for Infectious Diseases: Australian Capital Territory: Peter Collignon; The Canberra Hospital. New South Wales: Richard Benn (Royal Prince Alfred Hospital); Ian Chambers (Douglass Hanly Moir Pathology); Sharon Chen (Westmead Hospital); Nelson Dennis (Wollongong Hospital); Deo DeWit (Gosford Hospital); John Ferguson (John Hunter Hospital); Iain Gosbell (Liverpool Hospital); Thomas Gottlieb (Concord Hospital); Catriona Halliday (Westmead Hospital); Juliette Holland (Mayne Laverty Pathology); Alison Kesson (New Children's Hospital, Westmead); Richard Lawrence (St. George Hospital); Deborah Marriott (St. Vincent's Hospital, Sydney); Wieland Meyer (Westmead Hospital); Peter Newton (Wollongong Hospital); Quoc Nguyen (St. Vincent's Hospital, Sydney); Pamela Palasanthrian (Sydney Children's Hospital); Robert Pickles (infectious diseases physician, Taree), Robert Pritchard (Royal North Shore Hospital); Tania Sorrell (Westmead Hospital); Lex Tierney (John Hunter Hospital); Voula Tomasotos (Liverpool Hospital); Robert Vaz (Orange Base Hospital); Kerry Weeks (Royal North Shore Hospital). Queensland: Anthony Allworth (Royal Brisbane Hospital); Christopher Coulter (The Prince Charles Hospital); Joan Faoagali (Royal Brisbane Hospital); Barbara Johnson (Princess Alexandra Hospital), David Looke (Princess Alexandra Hospital), Joseph McCormack (The Mater Adult Hospital); Graeme Nimmo (Princess Alexandra Hospital); Gabrielle O'Kane (The Prince Charles Hospital); E. Geoffrey Playford (Princess Alexandra Hospital); Jennifer Robson (Sullivan and Nicolaides Pathology). South Australia: David Ellis (Women's and Children's Hospital); Rosemary Handke (Women's and Children's Hospital); Karen Rowlands (Royal Adelaide Hospital); David Shaw (Royal Adelaide Hospital). Tasmania: Louise Cooley (Royal Hobart Hospital); Erica Cox (Launceston General Hospital); Alistair McGregor (Royal Hobart Hospital). Victoria: Clare Franklin (Alfred Hospital); Cathy Joseph (St Vincent's Hospital, Melbourne), Tony Korman (Monash Medical Centre), Orla Morrissey (Alfred Hospital), Monica Slavin (Peter MacCallum Cancer Centre), Denis Spelman (Alfred Hospital); Bryan Speed (Austin and Repatriation Hospitals); Harsha Sheorey (St. Vincent's Hospital, Melbourne). Western Australia: Peter Boan (Royal Perth Hospital); John Dyer (Fremantle Hospital); Christopher Heath (Royal Perth Hospital); Dianne Gardam (Royal Perth Hospital); Duncan McLennan (Fremantle Hospital); Ronan Murray (Royal Perth Hospital); Todd Pryce (Royal Perth Hospital).
Please use the form below to submit correspondence to the authors or contact them at the following address:
Wieland Meyer, Molecular Mycology Research Laboratory, Westmead Hospital, Centre for Infectious Diseases and Microbiology, Institute of Clinical Pathology and Medical Research, Level 3, Room 3114A, Darcy Rd, Westmead, New South Wales 2145, Australia; email: w.meyer@usyd.edu.au
Please contact the EID Editors at eideditor@cdc.gov
The opinions expressed by authors contributing to this journal do not necessarily reflect the opinions of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
This page posted January 29, 2008
This page last reviewed March 20, 2008
Centers for Disease Control and Prevention, 1600 Clifton Rd, Atlanta, GA 30333, U.S.A
Tel: (404) 639-3311 / Public Inquiries: (404) 639-3534 / (800) 311-3435