|
Research
Risk to Human Health
from a Plethora of Simian Immunodeficiency Viruses in Primate Bushmeat
Martine Peeters,* Valerie Courgnaud,* Bernadette Abela,† Philippe
Auzel,†‡ Xavier Pourrut,* Frederic Bibollet-Ruche,§ Severin Loul,†
Florian Liegeois,* Cristelle Butel,* Denis Koulagna,¶ Eitel Mpoudi-Ngole,†
George M. Shaw,§ Beatrice H. Hahn,§ and Eric Delaporte*
*Institut de Recherche pour le Développement (IRD), Montpellier,
France; †Projet Prevention du Sida au Cameroun (PRESICA), Yaoundé,
Cameroon; ‡Faculté Universitaire des Sciences Agronomiques
de Gembloux, Gembloux, Belgium; §University of Alabama at Birmingham,
Birmingham, Alabama, USA; and ¶Ministry of Environment and Forestry,
Yaounde, Cameroon
To assess
human exposure to Simian immunodeficiency virus (SIV) in
west central Africa, we looked for SIV infection in 788 monkeys
that were hunted in the rainforests of Cameroon for bushmeat or
kept as pets. Serologic reactivity suggesting SIV infection was
found in 13 of 16 primate species, including 4 not previously
known to harbor SIV. Overall, 131 sera (16.6%) reacted strongly
and an additional 34 (4.3%) reacted weakly with HIV antigens.
Molecular analysis identified five new phylogenetic SIV lineages.
These data document for the first time that a substantial proportion
of wild monkeys in Cameroon are SIV infected and that humans who
hunt and handle bushmeat are exposed to a plethora of genetically
highly divergent viruses.
First recognized in the early 1980s, AIDS represents the endstage
of infection with one of two lentiviruses, termed Human immunodeficiency
virus type 1 (HIV-1) or type 2 (HIV-2) (1,2).
HIV-1 has spread to most parts of the world, while HIV-2 has remained
largely restricted to West Africa (3,4).
More than 40 million persons are estimated to have HIV infection
or AIDS (4).
Both HIV-1 and HIV-2 are of zoonotic origin (5).
The closest simian relatives of HIV-1 and HIV-2 have been found
in the common chimpanzee (Pan troglodytes) and the sooty
mangabey (Cercocebus atys), respectively (6-8),
and phylogenetic evidence indicates that lentiviruses from these
species (SIVcpz and SIVsm, respectively) have been transmitted to
humans on at least eight occasions (5,9).
Serologic evidence of SIV infection has so far been documented in
26 primate species, and 20 of these viruses have been at least partially
molecularly characterized (5,10,11).
Because humans come in frequent contact with primates in many parts
of sub-Saharan Africa, additional zoonotic transfers of primate
lentiviruses from species other than chimpanzees and sooty mangabeys
are possible. The risk for acquiring SIV infection would be expected
to be highest in persons who hunt primates and prepare their meat
for consumption, as well as in persons who keep primates as pets.
However, this risk cannot be assessed since the prevalence, diversity,
and geographic distribution of SIV infections in wild primate populations
are unknown. We report the first comprehensive survey of wild-caught
primates in Cameroon, home to diverse primate species that are extensively
hunted for food and trade (12). Much of the primate
meat sold for consumption derives from infected monkeys, and a comparable
number of pet monkeys also carry SIV. These data thus provide a
first approximation of the magnitude and variety of SIVs to which
humans are exposed through contact with nonhuman primates.
Materials
and Methods
Collection of Primate
Tissue and Blood Samples
Blood was obtained from 788 monkeys wild-caught in Cameroon from
January 1999 to April 2001. Species were determined by visual inspection
according to the Kingdon Field Guide to African Mammals
(13) and the taxonomy described by Colin
Groves (14). We sampled 573 animals
as bushmeat at markets in Yaoundé (n=157), surrounding villages
(n=111), or logging concessions in southeastern Cameroon (n=305),
as well as 215 pet animals from these same areas (Table
1). All primate samples were obtained with government approval
from the Cameroonian Ministry of Environment and Forestry. Bushmeat
samples were obtained through a strategy specifically designed not
to increase demand: women preparing and preserving the meat for
subsequent sale and hunters already involved in the trade were asked
for permission to sample blood and tissues from carcasses, which
were then returned.
For the bushmeat animals, blood was collected by cardiac puncture,
and lymph node and spleen tissues were collected whenever possible.
The owners indicated that most of the animals had died 12 to 72
hours before sampling. For pet monkeys, blood was drawn by peripheral
venipuncture after the animals were tranquilized with ketamine (10
mg/kg). Plasma and cells were separated on site by Ficoll gradient
centrifugation. All samples, including peripheral blood mononuclear
cells (PBMCs), plasma, whole blood, and other tissues, were stored
at –20°C.
Serologic Testing
Plasma samples were tested for HIV/SIV antibodies by the INNO-LIA
HIV Confirmation test (Innogenetics, Ghent, Belgium), which includes
HIV-1 and HIV-2 recombinant proteins and synthetic peptides that
are coated as discrete lines on a nylon strip. Five HIV-1 antigens
include synthetic peptides for the exterior envelope glycoprotein
(sgp120), as well as recombinant proteins for the transmembrane
envelope glycoprotein (gp41), integrase (p31), core (p24), and matrix
(p17) proteins. HIV-1 group O envelope peptides are included in
the HIV-1 sgp120 band. The HIV-2 antigens include synthetic peptides
for sgp120, as well as recombinant gp36 protein. In addition to
these HIV antigens, each strip has control lines: one sample addition
line (3+) containing anti-human immunoglobulin (Ig) and two test
performance lines (1+ and +/-) containing human IgG. All assays
were performed according to manufacturer’s instructions, with alkaline
phosphatase-labeled goat anti-human IgG as the secondary antibody.
We used the following working definition for SIV seropositivity:
plasma samples were scored as INNO-LIA positive when they reacted
with at least one HIV antigen and had a band intensity equal to
or greater than the assay cutoff (+/-) lane; samples that reacted
less strongly but still visibly with two or more HIV antigens were
classified as indeterminant; and samples reacting with no bands
or only one band with less than +/- intensity were classified as
negative.
Polymerase Chain Reaction
(PCR)
DNA was isolated from whole blood or PBMCs by using Qiagen DNA
extraction kits (Qiagen, Courtaboeuf, France), and PCR was done
with the Expand High Fidelity PCR kit (Roche Molecular Biochemicals,
Mannheim, Germany). For amplification of SIV sequences, previously
described degenerate consensus pol primers DR1, Polis4, UNIPOL2,
and PolOR (15-17) were used in various
combinations under previously described PCR conditions
(16). PCR products were sequenced by cycle sequencing
and dye terminator methods (ABI PRISM Big Dye Terminator Cycle Sequencing
Ready Reaction kit with AmpliTaq FS DNA polymerase [PE Biosystems,
Warrington, England]) on an automated sequencer (ABI 373, Stretch
model; Applied Biosystems, Courtaboeuf, France) either directly
or after cloning into the pGEM-T vector (Promega, Charbonnieres,
France).
To test for DNA degradation, a 1,151-bp region of the glucose-6–phosphate
dehydrogenase (G6PD) gene was amplified with the primers GPD-F1
5'-CATTACCAGCTCCATGACCAGGAC-3'and GPD-R1 5'-GTGTTCCCAGGTGACCCTCTGGC-3'
in a single-round PCR reaction under the following conditions: 94°C
for 2 min, then 35 cycles at 94°C for 20 sec; 58°C for 30 sec, and
72°C for 1 min (18).
Phylogenetic Analyses
Newly derived SIV nucleotide sequences were aligned with reference
sequences from the Los Alamos HIV/SIV Sequence database by using
CLUSTAL W (19) with minor adjustments
for protein sequences. A phylogenetic tree was constructed by the
neighbor-joining method (20), and the reliability
of branching orders was tested by the bootstrap approach (21).
Sequence distances were calculated by Kimura’s two-parameter method
(22). SIV lineages were defined as clusters
of SIV sequences from the same primate species that grouped together
with significant (>80%) bootstrap values.
GenBank Accession Numbers
The new sequences have been deposited in GenBank under the following
accession numbers: SIVgsn-99CM-CN71 (AF478588), SIVgsn-99CM-CN7
(AF478589), SIVgsn-99CM-CN166 (AF478590), SIVmon-99CM-CML1 (AF478591),
SIVmus-01CM-S1239 (AF478592), SIVmus-01CM-S1085 (AF478593), SIVtal-00CM-271
(AF478594), SIVtal-00CM-266 (AF478595), SIVmnd2-99CM-54 (AF478596),
SIVmnd2-01CM-S109 (AF478597), SIVmnd2-00CM-S46 (AF478598), SIVmnd2-00CM-S6
(AF478599), SIVdeb-01CM-1083 (AF478600), SIVdeb-99CM-CN40 (AF478601),
SIVdeb-01CM-S1014 (AF478602), SIVdeb-99CM-CNE5 (AF478603), SIVdeb-01CM-1161
(AF478604), SIVdeb-99CM-CNE1 (AF478605), SIVcol-00CM-247 (AF478606),
SIVcol-00CM-243 (AF478607), and SIVcol-99CM-11 (AF478608).
Results
Prevalence Estimates
of SIV Infection in Bushmeat and Pet Monkey Samples
Previous studies of SIV infection have relied almost exclusively
on surveys of captive monkeys or apes that were either kept as pets
or housed at zoos, sanctuaries, or primate centers. While this approach
has led to the discovery of novel SIVs (23-29),
it has not provided information concerning SIV prevalence rates
in the wild. Most pet monkeys are acquired at a very young age,
often when their parents are killed by hunters. Two field studies
of wild African green monkeys have shown that seroprevalence rates
correlated with sexual maturity, suggesting transmission predominantly
by sexual routes (30,31).
SIV infection rates of captive monkeys may thus not accurately reflect
SIV prevalence rates in the wild.
|
Figure
1 |
|
|
 |
|
|
Click
to view enlarged image
Figure 1. Detection
of HIV-1/HIV-2 cross-reactive antibodies in sera from 11 primate
species by using a line immunoassay (INNO-LIA HIV Confirmation,
Innogenetics, Ghent, Belgium)....
|
|
|
|
|
|
Figure
2
|
|
|
 |
|
|
Click
to view enlarged image
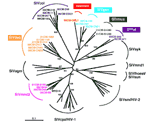
Figure 2. Identification
of diverse Simian immunodeficiency virus (SIV) lineages
in primate bushmeat.... |
|
To ensure systematic sampling, we therefore collected blood from
573 monkeys sold as bushmeat and 215 pet monkeys (Table
1). Most of the bushmeat animals were adults, while most of
the pets were still infants or juveniles at the time of sampling.
Most primates came from the southern part of the country. All major
SIV lineages known to date were initially discovered because their
primate hosts had antibodies that cross-reacted with HIV-1 or HIV-2
antigens (23-29). Although the extent of this
cross-reactivity has not been defined, we used a similar approach
to examine the primate blood samples obtained in Cameroon. Since
commercially available HIV screening assays (e.g., enzyme-linked
immunosorbent assay or rapid tests) contain only a limited number
of antigens, we used an HIV confirmatory assay (INNO-LIA), comprising
a recombinant and synthetic peptide-based line immunoassay (Figure
1). One hundred thirty-one (16.6%) of 788 plasma samples reacted
strongly with one or more HIV antigens, while an additional 34 samples
(4.3%) reacted less strongly but visibly with two or more HIV antigens
(Figure 1;Table 2).
Of 13 primate species that had HIV cross-reactive antibodies, the
prevalence of seroreactivity (positive plus indeterminant) ranged
from 5% to 40%. Prevalences were lower in pet animals than in bushmeat
primates, 11.6% versus 18.4%, respectively. Sera from only three
species failed to react completely (Cercopithecus preussi,
Mandrillus leucophaeus, Cercocebus torquatus), but these
three species accounted for only 5 of the 788 samples tested.
The INNO-LIA profiles from members of the same as well as different
primate species varied extensively (Figure 1).
Some sera reacted only with HIV core and/or Pol proteins, while
others reacted with Gag and/or Pol and/or Env proteins from either
HIV-1 or HIV-2 or both. Other than classifying sera as INNO-LIA
reactive or nonreactive, no banding pattern or algorithm could be
derived that would have been predictive of infection of any given
primate species.
Confirmation of SIV
Infection by PCR and Discovery of Novel SIV Lineages
A total of 342 samples, including INNO-LIA positive (n=91), indeterminant
(n=23), or negative (n=228) specimens were subjected to PCR analysis
(16,32), which yielded amplification
products for 28 blood samples from seven primate species: Cercopithecus
mona, C. neglectus, C. nictitans, C. cephus,
Colobus guereza, Miopithecus ogouensis, and Mandrillus sphinx
(Table 3). All these amplification products
were of appropriate size. Moreover, subsequent sequence and phylogenetic
analysis confirmed SIV infection (Figure 2).
Most of the newly derived sequences did not fall into any of the
known SIV groups. Viral sequences from C. mona (SIVmon),
C. neglectus (SIVdeb), C. nictitans (SIVgsn), C.
cephus (SIVmus), and Miopithecus ogouensis (SIVtal)
formed species-specific monophyletic clusters that were roughly
equidistant from each other as well as from all previously defined
SIV lineages in this region of the pol gene. Viruses from
the remaining two species (Colobus guereza and Mandrillus
sphinx) grouped with previously reported SIVcol and SIVmnd-2
strains, respectively.
The single sequence of SIVmon was given lineage status because
of its high degree of genetic diversity from the other SIV strains.
We maintained the lineage designation of SIVtal previously assigned
to a virus thought to be derived from a zoo animal of the species
M. talapoin (28) because that sequence
and the two newly derived talapoin viruses from M. ogouensis
cluster together in a phylogenetic tree derived from additional
pol nucleotide sequences (not shown). Thus, our new SIVtal
sequences confirm the existence of this lineage in the wild .
SIV sequences were confirmed in 26 of 91 INNO-LIA-positive samples,
as well as in 1 of 23 indeterminate and 1 of 223 negative samples
(Table 3). Because many blood samples were
obtained under poorly controlled circumstances, especially from
the bushmeat markets, we tested the possibility of DNA degradation.
Whole blood and PBMC DNA preparations were subjected to single-round
PCR with primers designed to amplify introns 4 and 5 of the nuclear
G6PD gene (1,100 bp). Of the 65 LIA-positive samples that did not
yield a virus-specific PCR product, 11 also failed to yield a G6PD
amplification product. Similarly, 4 of 17 INNO-LIA-indeterminate
and SIV PCR-negative samples, as well as 25 of 102 INNO-LIA-negative
samples, were also negative by G6PD amplification. These results
indicate that, in addition to using only a single set of nested
pol primer pairs, low PCR amplification rates from LIA-positive
and -indeterminant samples were also due to DNA degradation, the
presence of PCR inhibitors, or both.
Zoonotic transfers of SIV to humans have been documented on no
fewer than eight occasions (5,9), but no previous
study has examined to what extent African primates that are frequently
hunted or kept as pets are infected with SIV. Although our serologic
screening approach has limitations (i.e., an unknown extent of antigenic
cross-reactivity between HIV proteins and SIV antibodies), we were
able to detect cross-reactive antibodies suggesting SIV infection
in 16.6% of all tested animals, including members of four species
not previously known to harbor SIV (C. agilis, Lophocebus
albigena, C. pogonias, and Papio anubis). PCR
confirmation and molecular identification of SIV infection were
obtained in seven species, and phylogenetic analyses showed the
presence of highly divergent viruses that grouped according to their
species of origin. Four of these SIV lineages from mona (C. mona),
De Brazza’s (C. neglectus), mustached (C. cephus),
and greater spot-nosed (C. nictitans) monkeys have not previously
been recognized. Finally, we confirmed the SIVtal infection of wild
talapoin monkeys (Miopithecus ogouensis). These data establish
for the first time that a considerable proportion of wild-living
primates in Cameroon are infected with SIV, posing a potential source
of infection to those who come in contact with them. Our findings
bring to 30 the number of African nonhuman primate species known
or strongly suspected to harbor primate lentiviruses (5).
Our data likely still underestimate the prevalence and diversity
of naturally occurring SIV infections in Cameroon. First, not all
native primate species were tested, and many were undersampled because
they were either rare in the regions of Cameroon where we sampled
for this study or too small to be regularly hunted. For example,
the absence of reactive sera from drills and red-capped mangabeys,
two species known to harbor SIV (15,23),
must be due to the low number of blood samples (5/788) analyzed.
In addition, the INNO-LIA test sensitivity is clearly not 100%,
as one negative sample contained SIV sequences as determined by
PCR amplification. Finally, our PCR approach, which utilized only
a single set of nested primers, likely amplified only a subset of
viral sequences. Thus, the true prevalence of SIV infection in the
various primate species will require the development of SIV lineage-specific
assays with known sensitivities and specificities.
Human infection with SIVcpz and SIVsm is thought to have resulted
from cutaneous or mucous membrane exposure to infected blood during
the hunting and butchering of chimpanzees and sooty mangabeys for
food (5). Bites from pet animals and possibly contact
with fecal and urine samples may have also been involved (5).
Our study shows that many primate species in addition to chimpanzees
and sooty mangabeys are hunted and that 20% (or more) of these animals
likely harbor SIV. Thus, if contact with infected blood or other
secretions is indeed the primary route of transmission, hunters
and food handlers may be at risk of infection with many more SIVs
than just those from chimpanzees and sooty mangabeys.
Bushmeat hunting, to provide animal proteins for the family and
as a source of income, has been a longstanding common component
of household economies in the Congo Basin and, more generally, throughout
subSaharan Africa (33-35). However, the bushmeat
trade has increased in the last decades. Commercial logging, which
represents an important economic activity in Cameroon as well as
many other west-central African countries, has led to road constructions
into remote forest areas, human migration, and social and economic
networks supporting this industry (36). Hunters
are now penetrating previously inaccessible forest areas, making
use of newly developed infrastructure to capture and transport bushmeat
from remote areas to major city markets (37).
Moreover, villages around logging concessions have grown from a
few hundred to several thousand inhabitants in just a few years
(37). These socioeconomic changes, combined with
our estimates of SIV prevalence and genetic complexity in wild primates,
suggest that the magnitude of human exposure to SIV has increased,
as have the social and environmental conditions that would be expected
to support the emergence of new zoonotic infections.
Whether any of the newly identified SIVs have the ability to infect
humans remains unknown since molecular evidence is lacking for SIV
cross-species transmissions from primates other than chimpanzees
and sooty mangabeys. However, such infections may have been unrecognized
by HIV-1/HIV-2 screening assays. A case in point is the recent identification
of a Cameroonian man who had an indeterminant HIV serology but reacted
strongly (and exclusively) with an SIVmnd V3 loop peptide (32).
Although viral sequences were not confirmed in this man, the finding
suggests that at least some naturally occurring SIVs have the potential
to cross the species into the human population. In fact, several
recently reported SIV isolates, including SIVlhoest, SIVsun, SIVrcm,
and SIVmnd2, replicate well in primary human lymphocytes in vitro
(23,26,27,32,38) as
do SIVcpz (25) and SIVsm (24).
Thus, to determine whether additional zoonotic transmissions of
SIVs have already occurred, virus type- and/or lineage-specific
immunoassays and PCRs will have to be developed. Such work should
receive high priority given the extent of human exposure to different
SIV lineages as a result of the expanding bushmeat trade and the
impact of two major human zoonoses (HIV-1 and HIV-2). Recombination
between newly introduced SIVs and circulating HIVs poses still another
human risk for novel zoonoses.
In summary, the current HIV-1 pandemic provides compelling evidence
for the rapidity, stealth, and clinical impact that can be associated
with even a single primate lentiviral zoonotic transmission event.
We document for the first time that humans are exposed to a plethora
of primate lentiviruses through hunting and handling of bushmeat
in Cameroon, a country at the center of HIV-1 groups M, N, and O
endemicity that is home to a diverse set of SIV-infected nonhuman
primates. To what extent wild monkey populations in other parts
of Africa are also infected with diverse SIVs is unknown. A complete
and accurate assessment of all SIV-infected nonhuman primate species
is needed, as well as a determination of the virus lineage(s) present
in each species. Studies are also needed to determine whether zoonotic
transmissions of SIVs from primates other than chimpanzees and mangabeys
have already occurred and what clinical outcomes were associated
with these infections. Results from these studies will yield critical
insights into the circumstances and factors that govern SIV cross-species
transmission and thus allow determination of human zoonotic risk
for acquiring these viruses.
Acknowledgments
We thank the Cameroonian Ministries of Health, Environment and
Forestry for permission to perform this study, the staff from the
PRESICA project for logistical support and assistance in the field,
and Caroline Tutin for scientific discussions.
This work was supported in part by grants from the Agence National
de Recherche sur le SIDA (ANRS) and the National Institutes of Health
(RO1 AI 44596, RO1 AI 50529, N01 AI85338, P30 AI 27767).
Dr. Peeters is director of research at the Institute for Research
and Development (IRD), Montpellier, France. Her major interests
are the molecular biology and epidemiology of human and simian immunodeficiency
viruses.
References
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre
MT, Chamaret S, Gruest J, et al. Isolation
of a T-lymphotropic retrovirus from a patient at risk for acquired
immune deficiency syndrome (AIDS). Science 1983;220:868-70.
- Clavel F, Mansinho K, Chamaret S, Guetard D, Favier V, Nina
J, et al. Human
immunodeficiency virus type 2 infection associated with AIDS in
West Africa. N Engl J Med 1987:316;1180-5.
- UNAIDS. 2000. Report on the global HIV/AIDS epidemic. Available
at: URL: http://www.unaids.org/
- van der Loeff MFS, Aaby P. Towards
a better understanding of the epidemiology of HIV-2. AIDS
1999;13:S69-S84.
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS
as a zoonosis: scientific and public health implications.
Science 2000;287:607-17.
- Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S.
Genetic organization of a chimpanzee lentivirus related to HIV-1.
Nature 1990;345:356-9.
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael
SF, et al.
Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes.
Nature 1999;397:436-41.
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR.
An
African primate lentivirus (SIVsm) closely related to HIV-2.
Nature 1989;339:389-92.
- Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO,
Hahn BH. The
origins of AIDS viruses: where and when? Philos Trans R Soc
Lond B Biol Sci 2001;356:867-6.
- Lowenstine LJ, Pedersen NC, Higgins J, Pallis KC, Uyeda A, Marx
P, et al. Seroepidemiologic
survey of captive Old World primates for antibodies to human and
simian retroviruses, and isolation of a lentivirus from sooty
mangabeys (Cercocebus atys). Int J Cancer 1986;38:563-74.
- Nicol I, Messinger D, Dubouch P, Bernard J,
Desportes I, Jouffre R, et al. Use
of Old World monkeys for acquired immunodeficiency syndrome research.
J Med Primatol 1989;18:227-36.
- Bennett EL, Robinson JG. Hunting for the snark. In: Robinson
JG, Bennett EL, editors. Hunting for sustainability in tropical
forests. New York: Columbia University Press; 2000. p. 1-9.
- Kingdon J. The Kingdon field guide to African mammals. San Diego:
Academic Press; 1997.
- Groves C. Primate taxonomy. Washington: Smithsonian Institution
Press; 2001.
- Clewley JP, Lewis JCM, Brown DWG, Gadsby EL. Novel
simian immunodeficiency virus (SIVdrl) pol sequence from the drill
monkey, Mandrillus leucophaeus. J Virol 1998;72:10305-9.
- Courgnaud V, Pourrut X., Bibollet-Ruche F, Mpoudi-Ngole E, Bourgeois
A, Delaporte E, et al. Characterization
of a novel simian immunodeficiency virus from Guereza Colobus
(Colobus guereza) in Cameroon: a new lineage in the nonhuman
primate lentivirus family. J Virol 2001;75:857-66.
- Miura T, Sakuragi J, Kawamura M, Fukasawa M, Moriyama EN, Gojobori
T, et al. Establishment
of a phylogenetic survey system for AIDS-related lentiviruses
and demonstration of a new HIV-2 subgroup. AIDS 1990;4:1257-61.
- von Dornum M, Ruvolo M.
Phylogenetic relationships of the new world monkeys (Primates,
Platyrrhini) based on nuclear G6PD DNA sequences. Mol Phylogenet
Evol 1999;11:459-76.
- Thompson JD, Higgins DG, Gibson TJ.
CLUSTAL W - improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific
gap penalties and weight matrix choice. Nucleic Acids Res
1994;22:4673-80.
- Saitou N, Nei M.
The neighbor-joining method: a new method for reconstructing phylogenetic
trees. Mol Biol Evol 1987;4:406-25.
- Felsenstein J. Confidence-limits on phylogenies—an
approach using the bootstrap. Evolution 1985;39:783-91.
- Kimura M. The neutral theory of molecular evolution. Cambridge:
Cambridge University Press; 1983.
- Georges-Courbot MC, Lu CY, Makuwa M, Telfer P, Onanga R, Dubreuil
G, et al.
Natural infection of a household pet red-capped mangabey (Cercocebus
torquatus torquatus) with a new simian immunodeficiency virus.
J Virol 1998;72:600-8.
- Peeters M, Janssens W, Fransen K, Brandful J, Heyndrickx L,
Koffi K, et al.
Isolation of simian immunodeficiency viruses from two sooty mangabeys
in Cote d'Ivoire: virological and genetic characterization and
relationship to other HIV type 2 and SIVsm/mac strains. AIDS
Res Hum Retroviruses 1994;10:1289-94.
- Peeters M, Fransen K, Delaporte E, Van den Haesevelde M, Gershy-Damet
GM, Kestens L, et al. Isolation
and characterization of a new chimpanzee lentivirus (simian immunodeficiency
virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS
1992;6:447-51.
- Beer BE, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley
SG, et al. Simian
immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus
solatus): evidence for host-dependent evolution of SIV within
the C. lhoesti superspecies. J Virol
1999;73:7734-44.
- Hirsch VM, Campbell BJ, Bailes E, Goeken R, Brown C, Elkins
WR, et al. Characterization
of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys
(Cercopithecus l'hoesti): implications for the origins
of SIVmnd and other primate lentiviruses. J Virol 1999;73:1036-45.
- Osterhaus AD, Pedersen N, van Amerongen G, Frankenhuis MT, Marthas
M, Reay E, et al.
Isolation and partial characterization of a lentivirus from Talapoin
Monkeys (Myopithecus talapoin). Virology 1999;260:116-24.
- Tsujimoto H, Hasegawa A, Maki N, Fukasawa M, Miura T, Speidel
S, et al. Sequence
of a novel simian immunodeficiency virus from a wild-caught African
mandrill. Nature 1989;341:539-41.
- Phillips-Conroy JE, Jolly CJ, Petros B, Allan JS, Desrosiers
RC.
Sexual transmission of SIVagm in wild grivet monkeys. J Med
Primatol 1994;23:1-7.
- Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado
P, Galat G, Durand JP, et al. Simian
immunodeficiency virus infection in a patas monkey (Erythrocebus
patas): evidence for cross-species transmission from African
green monkeys (Cercopithecus aethiops sabaeus) in the wild.
J Gen Virol 1996;77:773-81.
- Souquiere S, Bibollet-Ruche F, Robertson DL, Makuwa M, Apetrei
C, Onanga R, et al. Wild
Mandrillus sphinx are carriers of two types of lentiviruses.
J Virol 2001;75:7086-96.
- Asibey
EO. Wildlife as a source of protein in Africa south of the Sahara.
Biol Conserv 1974;6:32-9.
- Geist V. How markets for wildlife meat and parts, and the sale
of hunting privileges, jeopardize wildlife conservation. Conser
Biol 1988;2:15-26.
- Chardonnet P. Faune sauvage africaine: la resource oubliee.
Fondation Internationale Pour la Sauvegarde de la nature/CIRAD-EMVT.
Luxembourg: Office des publications officielles des communautés
européennes;1996.
- Wilkie D, Shaw E, Rotberg F, Morelli G, Auzel P. Roads, development,
and conservation in the congo Basin. Conser Biol 2000;14:1614-22.
- Auzel P, Hardin R. Colonial history, concessionary politics,
and collaborative management of Equatorial African rain forests.
In: Bakarr M, Da Fonseca G, Konstant W, Mittermeier R, Painemilla
K, editors. Hunting and bushmeat utilization in the African rain
forest. Washington: Conservation International; 2000. p 21-38.
- Beer BE, Foley BT, Kuiken CL, Tooze Z, Goeken RM, Brown CR,
et al. Characterization
of novel simian immunodeficiency viruses from red-capped mangabeys
from Nigeria (SIVrcmNG409 and -NG411). J Virol 2001;75:12014-27.
| Table
1.Wild-born primates surveyed, by species, age, and status,
Cameroon |
|
| Genus |
Species |
Common name |
Pet animals
|
Primate bushmeat
|
Total
|
|
|
|
|
Adults
|
Juveniles/Infants
|
Adults
|
Juveniles/infants
|
|
| Cercocebus |
agilis
|
Agile mangabey |
4
|
15
|
30
|
3
|
52
|
| torquatus |
Red-capped
mangabey |
1
|
–
|
–
|
1
|
2
|
|
Lophocebus
|
albigena
|
Grey-cheeked mangabey
|
3
|
3
|
12
|
3
|
21
|
|
Cercopithecus
|
cephus
|
Mustached guenon
|
3
|
26
|
217
|
56
|
302
|
|
mona
|
Mona monkey
|
–
|
7
|
1
|
1
|
9
|
|
neglectus
|
De Brazza’s monkey
|
2
|
6
|
21
|
5
|
34
|
|
nictitans
|
Greater spot-nosed monkey
|
8
|
36
|
110
|
12
|
166
|
|
pogonias
|
Crested mona
|
1
|
5
|
57
|
10
|
73
|
|
preussi
|
Preuss’s monkey
|
–
|
1
|
–
|
–
|
1
|
|
Chlorocebus
|
tantalus
|
Tantalus monkey
|
7
|
11
|
–
|
–
|
18
|
|
Miopithecus
|
ogouensis
|
Gabon talapoin
|
5
|
6
|
8
|
–
|
19
|
|
Erytrocebus
|
patas
|
Patas monkey
|
5
|
14
|
–
|
–
|
19
|
|
Colobus
|
guereza
|
Mantled guereza
|
–
|
2
|
24
|
–
|
26
|
|
Mandrillus
|
leucophaeus
|
Drill
|
–
|
2
|
–
|
–
|
2
|
|
sphinx
|
Mandrill
|
5
|
15
|
–
|
2
|
22
|
|
Papio
|
anubis
|
Olive baboon
|
11
|
11
|
–
|
–
|
22
|
|
Total
|
|
|
55
|
160
|
480
|
93
|
788
|
|
|
|
| Table
2. HIV-1/HIV-2 cross-reactive antibodiesa detected
in primate species, Cameroon |
|
| Genus |
Species |
Common name |
Pet animals
|
Primate bushmeat
|
Total
|
|
|
|
|
pos/tested
|
ind/tested
|
pos/tested
|
ind/tested
|
pos/tested
|
ind/tested
|
|
| Cercocebus |
agilis
|
Agile mangabey
|
1/19
|
1/19
|
5/33
|
7/33
|
6/52
|
8/52
|
|
torquatus
|
Red-capped mangabey
|
0/1
|
0/1
|
0/1
|
0/1
|
0/2
|
0/2
|
|
Lophocebus
|
albigena
|
Grey-cheeked mangabey
|
0/6
|
0/6
|
2/15
|
3/15
|
2/21
|
3/21
|
|
Cercopithecus
|
cephus
|
Mustached guenon
|
1/29
|
3/29
|
48/273
|
9/273
|
49/302
|
12/302
|
|
mona
|
Mona monkey
|
1/7
|
0/7
|
1/2
|
0/2
|
2/9
|
0/9
|
|
neglectus
|
De Brazza’s monkey
|
1/8
|
0/8
|
9/26
|
1/26
|
10/34
|
1/34
|
|
nictitans
|
Greater spot-nosed monkey
|
6/44
|
0/44
|
22/122
|
3/122
|
28/166
|
3/166
|
|
pogonias
|
Crested mona
|
0/6
|
0/6
|
9/67
|
4/67
|
9/73
|
4/73
|
|
preussi
|
Preuss’s monkey
|
0/1
|
-
|
-
|
-
|
0/1
|
-
|
|
Chlorocebus
|
tantalus
|
Tantalus monkey
|
3/18
|
0/18
|
-
|
-
|
3/18
|
0/18
|
|
Miopithecus
|
ogouensis
|
Gabon talapoin
|
2/11
|
1/11
|
2/8
|
0/8
|
4/19
|
1/19
|
|
Erythrocebus
|
patas
|
Patas monkey
|
1/19
|
0/19
|
-
|
-
|
1/19
|
0/19
|
|
Colobus
|
guereza
|
Mantled guereza
|
0/2
|
0/2
|
7/24
|
1/24
|
7/26
|
1/26
|
|
Mandrillus
|
leucophaeus
|
Drill
|
0/2
|
0/2
|
-
|
-
|
0/2
|
0/2
|
|
sphinx
|
Mandrill
|
7/20
|
0/20
|
1/2
|
1/2
|
8/22
|
1/22
|
|
Papio
|
anubis
|
Olive baboon
|
2/22
|
0/22
|
-
|
-
|
2/22
|
0/22
|
|
Total
|
|
|
25/215
|
5/215
|
106/573
|
29/573
|
131/788
|
34/788
|
|
(%)
|
|
|
11.6
|
2.3
|
18.4
|
5.1
|
16.6
|
4.3
|
|
|
aPlasma samples were tested for
antibodies cross-reactive with HIV-1 and HIV-2 antigens by
using a recombinant-based line immunoassay (INNO-LIA HIV Confirmation,
Innogenetics, Ghent, Belgium). Positive (pos) and indeterminant
(ind) INNO-LIA scoring criteria as described in Methods.
|
| Table
3. Polymerase chain reaction (PCR) amplification of Simian
immunodeficiency virus (SIV) sequences |
|
|
Genus
|
Species
|
INNO-LIA posa
PCR pos/tested
|
INNO-LIA ind
PCR pos/tested
|
INNO-LIA neg
PCR pos/tested
|
|
|
Cercocebus
|
agilis
|
0/6
|
0/8
|
0/13
|
|
torquatus
|
–
|
–
|
0/1
|
|
Lophocebus
|
albigena
|
0/2
|
0/2
|
0/7
|
|
Cercopithecus
|
cephus
|
2/25
|
0/7
|
0/56
|
|
mona
|
1/2
|
–
|
0/2
|
|
neglectus
|
8/9
|
–
|
0/4
|
|
nictitans
|
3/21
|
1/1
|
0/61
|
|
pogonias
|
0/9
|
0/3
|
0/34
|
|
Chlorocebus
|
tantalus
|
0/1
|
–
|
0/2
|
|
Miopithecus
|
ogouensis
|
2/3
|
–
|
0/10
|
|
Erythrocebus
|
patas
|
–
|
–
|
0/7
|
|
Colobus
|
guereza
|
6/6
|
0/1
|
1/16
|
|
Mandrillus
|
sphinx
|
4/5
|
0/1
|
0/4
|
|
Papio
|
anubis
|
0/2
|
–
|
0/11
|
| Total |
|
26/91
|
1/23
|
1/228
|
|
|
aDNA was extracted from a subset
of seropositive (pos), indeterminant (ind) and negative (neg)
blood samples and subjected to nested PCR amplification by
using HIV/SIV consensus pol primer pairs. In each column,
the number of PCR-positive samples per total number of samples
tested is indicated. The authenticity of all amplification
products was confirmed by sequence analysis.
|
|
