|
U.S. Pandemic Flu Model 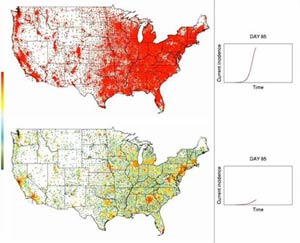
These simulations show the hypothetical spread of a moderately contagious pandemic flu in the United States. Each dot represents a Census tract and changes color from green to red as more people in that tract become infected. The dots change back to green as people recover. With no intervention (top), the pandemic peaks around day 85. With the distribution of 10 million doses per week of a vaccine that’s poorly matched to the emerging virus (bottom), the pandemic peaks around day 108.
View with RealPlayer or Windows Media Player.
All graphics are courtesy of the Proceedings of the National Academy of Sciences. |
If pandemic flu were to emerge in the United States, what interventions might slow its spread and minimize the impact? With support from the National Institutes of Health (NIH), researchers from the Fred Hutchinson Cancer Research Center in Seattle, Wash., and the Los Alamos National Laboratory have used computer models to suggest possible answers. The findings appear in the April 11, 2006, issue of the Proceedings of the National Academy of Sciences and will be available in the online edition the week of April 3.
By developing a model that represents the U.S. population and tests different properties of a potential pandemic flu virus, the researchers evaluated the effectiveness of different intervention strategies. They found that, depending on the contagiousness of the virus, a variety of approaches could reduce the number of cases to less than that of an annual flu season.
“Preparing for a potential pandemic is tremendously challenging, given the potential scope and the large number of unknowns,” said NIH Director Elias A. Zerhouni, M.D. “The best approach is to use all of the tools available to us, including computer modeling. By predicting the impact of intervention strategies, these models can help health officials and policymakers plan for a real pandemic.”
The recent modeling work is part of an ongoing research program called the Models of Infectious Disease Agent Study (MIDAS), supported by NIH’s National Institute of General Medical Sciences (NIGMS). Researchers in the network develop computer models to better understand the spread of infectious diseases, whether they occur naturally or deliberately. With growing concerns that the H5N1 strain of the avian flu virus, initially found in birds throughout Southeast Asia, could eventually be transmitted easily between people, the research network has been modeling pandemic flu in different parts of the world.
“The MIDAS researchers previously developed models of a potential pandemic flu outbreak in Thailand and surrounding areas that showed containment at the source is feasible,” explained Jeremy M. Berg, Ph.D., NIGMS director. “But we need to consider the possibility that if the outbreak isn’t contained, it could quickly spread globally.”
Using data from the 2000 U.S. Census and the U.S. Department of Transportation, the researchers developed a model that represents the demographics and travel patterns of 281 million people living in the United States. They also incorporated information about the potential virus based on previous flu pandemics, including different assumptions about its possible contagiousness (but not its potential effects on mortality). The researchers then introduced a small number of hypothetical travelers, who are infected but not yet symptomatic, arriving daily at 14 major U.S. international airports. With these assumptions in place, the scientists simulated a virtual outbreak on high-performance computers at the Los Alamos National Laboratory.
“The goal for the U.S. modeling project was to determine how to slow spread long enough so that a well-matched vaccine could be developed and distributed,” said the research team’s leader, Ira M. Longini, Jr., Ph.D., a biostatistician at the Fred Hutchinson Cancer Research Center and the University of Washington School of Public Health and Community Medicine. An additional guideline was to reduce the number of overall cases to or below 10 percent of the population, the average percentage reported during an annual flu season.
To identify such measures, the researchers tested different interventions: distributing antiviral treatments to infected individuals and others near them to reduce symptoms and susceptibility; vaccinating people, possibly children first, with either one or two shots of a vaccine not well matched to the strain that may emerge; social distancing, such as restricting travel and quarantining households; and closing schools.
The results showed that with no intervention a pandemic flu with low contagiousness could peak after 117 days and infect about 33 percent of the U.S. population. A highly contagious virus could peak after 64 days and infect about 54 percent of people.
The researchers then compared what might happen in scenarios involving the use of different interventions. When the simulated virus was less contagious, the three most effective single measures included distributing several million courses of antiviral treatment to targeted groups seven days after a pandemic alert, school closures, and vaccinating 10 million people per week with one dose of a poorly matched vaccine. The results also showed that vaccinating school children first is more effective than random vaccination when the vaccine supply is limited. Regardless of contagiousness, social distancing measures alone had little effect.
But when the virus was highly contagious, all single intervention strategies left nearly half the population infected. In this instance, the only measures that reduced the number of cases to below the annual flu rate involved a combination of at least three different interventions, including a minimum of 182 million courses of antiviral treatment.
While the results are specific to the United States, the researchers said the general findings can apply to other developed countries and could aid the drafting of preparedness plans both here and abroad. Because computer models can’t capture all the complexities of real communities and outbreaks, MIDAS scientists will continue to refine their models and test different scenarios as new information becomes available.
Other members of the Longini team who contributed to the recent findings include Los Alamos National Laboratory scientists Timothy C. Germann, Ph.D.; Kai Kadau, Ph.D.; and Catherine A. Macken, Ph.D.
###
To arrange an interview with NIGMS Director Jeremy M. Berg, Ph.D., contact the NIGMS Office of Communications and Public Liaison at 301-496-7301. To arrange an interview with Ira M. Longini, Jr., Ph.D., please call 206-667-2896. More information about MIDAS is available at http://www.nigms.nih.gov/Initiatives/MIDAS/.
NIGMS (http://www.nigms.nih.gov/) is one of 27 components of the National Institutes of Health, U.S. Department of Health and Human Services. The NIGMS mission is to support basic biomedical research that lays the foundation for advances in disease diagnosis, treatment, and prevention.
The National Institutes of Health (NIH)--The Nation's Medical Research Agency--includes 27 Institutes and Centers and is a component of the U. S. Department of Health and Human Services. It is the primary Federal agency for conducting and supporting basic, clinical, and translational medical research, and it investigates the causes, treatments, and cures for both common and rare diseases. For more information about NIH and its programs, visit http://www.nih.gov.
