Antimicrobial Resistance
Table of Contents
- What is antimicrobial resistance?
- What are the mechanisms of resistance?
- Why has antimicrobial resistance become such a problem?
- Controlling the Spread of Resistance
- Antimicrobial Resistance in Alaska
- Projects in Progress
- References
What is antimicrobial resistance?
The discovery of effective agents to prevent and treat infections caused by bacteria and other pathogenic microorganisms has been one of the most important developments of modern medicine. However, microorganisms have a short generation time-from minutes to hours-and can therefore respond rapidly to changes in their environment. Thus, as antimicrobial agents are introduced into the environment, microorganisms respond to the selective pressures of these agents by becoming resistant—that is, able to survive and reproduce in the presence of the agent. And, as long as antimicrobial agents are in use, this process will continue to occur. The consequences of increasing resistance are enormous. Resistance leads to increased morbidity and mortality, the need to use more toxic and expensive therapies, and the need to expend ever greater resources on monitoring the development of resistance and surveillance.
What are the mechanisms of resistance?
Mechanisms of acquired resistance result from changes in the cellular physiology and structure of a microorganism due to alterations in its usual genetic makeup. There are three main mechanisms whereby bacteria initially susceptible to an antimicrobial agent may acquire the ability to resist the effects of an agent [1]. These include:
- Prevention of intracellular drug accumulation by:
- alterations in the bacterial outer membrane, such that the drug no longer binds to the exterior of the micro-organism
- active transport of the drug across the cytoplasmic membrane in the cell ceases, or
- active efflux (pumping) of the drug out of the cell before it can damage the micro-organism
- Alteration in the drug target (the part of the micro-organism where damage is done) which leads to ineffective levels of drug binding to or near the target site;
- Production by the microorganism of a drug-inactivating enzyme that destroys or greatly diminishes the drug's ability to kill the micro-organism.
Many of these mechanisms result from genetic mutations, the acquisition of resistance genes from other microorganisms via gene transfer, and combinations of these two types of events.
Why has antimicrobial resistance become such a problem?
The origins of the current crisis in antimicrobial resistance are complex and may differ somewhat for hospital and community-acquired pathogens and for developed and underdeveloped countries [2]. In much of the industrial world, antimicrobial drugs (often called "antibiotics") are available only by prescription. In many other countries, a prescription is not required for the purchase of antimicrobial agents. Whether or not a prescription is obtained however, does not guarantee that antimicrobial agents will be used properly. Taken for nonbacterial infections (such as the common cold), in ineffective doses, or for an inadequate or inappropriate length of time, antimicrobial agents cannot only be ineffective, but exert strong selective pressures upon bacterial populations, favoring those organisms that are capable of resisting them. In addition to human uses of antimicrobial agents, there are a number of non-human applications that increases the pool of resistant organisms that may impact the clinical effectiveness of many antimicrobial agents used to treat infectious diseases of man. These applications include production of food animals (livestock, poultry), aquaculture, plant and crop protection, and food production [2,3].
Controlling the Spread of Resistance
Today, virtually all important bacterial infections in the U.S. and throughout the world are becoming resistant. For this reason, antimicrobial resistance is among CDC's top concerns. As a result, CDC has instituted a National Campaign for Appropriate Antibiotic Use which has two objectives [4]:
- Reduce inappropriate antibiotic use
- Reduce the spread of resistance to antibiotics
To accomplish these objectives, the campaign is:
- Developing and distributing educational materials promoting appropriate antibiotic use.
- Funding states to develop, implement and evaluate local campaigns.
- Developing and pilot testing a medical school curriculum to teach a new generation of physicians about the appropriate use of antibiotics.
- Funding a national advertising campaign promoting the appropriate use of antibiotics.
Antimicrobial Resistance in Alaska
Streptococcus pneumoniae
In Alaska, Streptococcus pneumoniae
is a major cause of pneumonia, but cases of meningitis, pericarditis, empyema,
septic arthritis, cellulitis, bacteremia and septic shock are also seen. Alaska
Natives have consistently had higher rates of invasive pneumococcal disease
than non-Natives in Alaska as well as other US populations. In addition to
high rates of disease, pneumococcal isolates with resistance to multiple classes
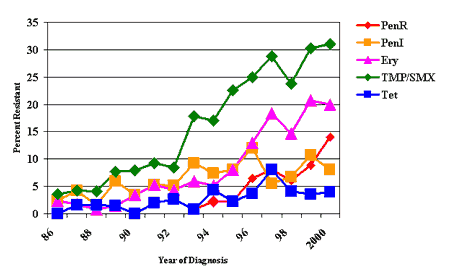
of antibiotics have been and continue to be recovered in Alaska [5][Fig.
1]. The percentage of invasive pneumococcal isolates which had any resistance
to penicillin increased from ~2% in 1986, to over 25% in recent years.
Isolates fully resistant to penicillin were first seen in Alaska in 1993 and
have steadily increased to nearly 15% in 2000. Resistance to other commonly
used antibiotics has also increased: erythromycin resistance increased from
~2% to 20% and trimethoprim/sulfamethoxazole increased from ~4%
to over 30%. The long-term solution for control of the spread of pneumococcal
isolates resistant to multiple classes of antibiotics is prevention. In Alaska
Natives and other populations with a high prevalence of infection, prevention
will include administering the 23-valent pneumococcal polysaccharide vaccine
to people who are at greatest risk of disease, use of efficacious conjugate
vaccines in young children, and measures to promote judicious use of antimicrobial
agents.
For more information see the Streptococcus
pneumoniae page.
Figure 1: Trends in antimicrobial resistance among invasive Pneumococcal isolates, Alaska, 1986-2000
Methicillin-resistant Staphylococcus aureus
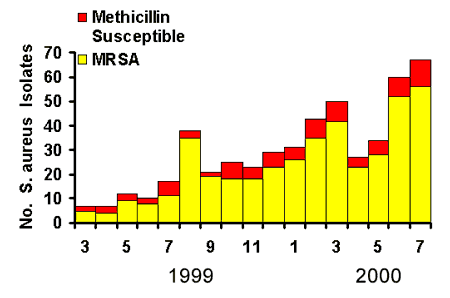
While infections due to S. aureus have long been common among rural Alaskans, infections due to methicillin-resistant S. aureus (MRSA) are a much more recent phenomenon presenting clinicians with significant therapeutic challenges. Evaluation of laboratory and medical records from a large outbreak of community-acquired MRSA skin and soft tissue infections in southwestern Alaska during 1999 and 2000 revealed that over 80% of culture-confirmed S. aureus infections were MRSA and 84% of the MRSA infections involved skin or soft tissues, with more serious and invasive disease being rare [6][Fig.2, Baggett, et al.] It was also found that patients with MRSA skin infections were more likely to have been prescribed antibiotics in the 180 days before onset of their infection. MRSA skin infections were also found to be more common among people who used crowded steam baths. Additionally, it has been found that MRSA isolates from rural Alaska are much more likely to carry the gene for a virulence factor associated with skin and soft tissue infections (the Panton/Valentine leukocidin) when compared with drug-susceptible strains. Prevention and control measures include:
- Use of appropriate antibiotics when needed
- Non-drug management of infections when possible
- Cleaning steam baths after every use
- Use of personal seating barriers in steam baths
- Education about handwashing and wound care
For more information see the methicillin-resistant S. aureus page.
Figure 2: S. aureus skin infection isolates, regional hospital, March 1999 - July 2000.
Helicobacter pylori
A serosurvey of over 2000 serum specimens collected in the 1980s from Alaska
Native communities revealed that 75% were positive for H.
pylori [7]. Rates increased from 32% among 0 to
4 years olds up to 86% in those 20 years or older. Rates also varied depending
on the region of the state, which was most pronounced among 0 to 4 year olds,
where rates ranged from 5% in southcentral to 65% in interior Alaska. Laboratory-based
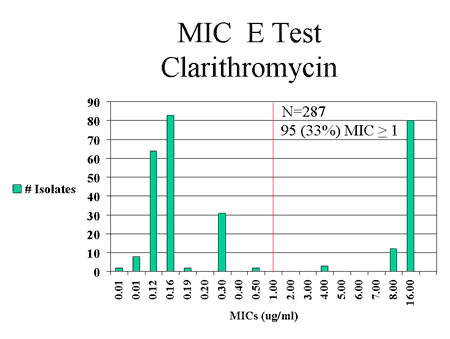
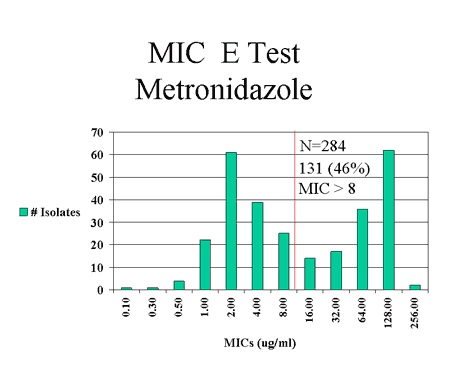
surveillance for H. pylori infections have found that a high proportion
of H. pylori isolates recovered from Alaska Native patients are resistant
to antibiotics commonly used in this population [8][Fig.
3]. Analysis of data from a prospective study of treatment outcomes for
Alaska Natives with H. pylori infection in Anchorage document that
infection with drug-resistant strains are more common in persons previously
treated with antibiotics and that resistance to clarithromycin is associated
with treatment failure.
For more information see the Helicobacter
pylori page.
Figure 3: Antimicrobial susceptibilities of H. pylori isolates recovered from Alaska Native patients at 3 Alaska hospital surveillance sites, July 1999 - July 2001.
Projects in Progress
- Ongoing surveillance for nasopharyngeal carriage of drug-resistant S. pneumoniae in rural Alaska villages to determine the impact of the decline in antimicrobial drug use on the prevalence of drug-resistant pneumococci in the community over time.
- Ongoing surveillance of S. pneumoniae in rural and urban Alaska to assess the impact of pneumococcal conjugate vaccines on drug-resistant S. pneumoniae disease and carriage.
- Studies of methods to reduce infections caused by MRSA in the community.
- Laboratory-based surveillance for H. pylori infections in Alaska Natives to monitor antimicrobial resistance.
- Examine reinfection with H. pylori after successful eradication to determine if antibiotic resistance is associated with past antibiotic usage and to determine if antimicrobial resistance patterns predicts treatment outcome.
References
1. Sanders CC, Sanders WE. Resistance to antibacterial
agents. In: Jungkind DI, Mortensen JE, Fraimow HS, Calandra GB, eds. Antimicrobial
Resistance, A Crisis in Health Care. Advances in Experimental Medicine
and Biology. Vol. 390. New York, NY: Plenum Press; 1995.
2. http://www.asm.org/Academy/index.asp?bid=2167
3. Shea KM. Antibiotic resistance: what is the impact of
agricultural uses of antibiotics on children's health? Pediatrics 2003;1(112):253-58.
4. http://www.cdc.gov/drugresistance/
5. Rudolph, KM, Parkinson AJ, Reasonover AL, Bulkow LR,
Parks DJ, Butler JC.
Serotype distribution and antimicrobial resistance patterns of invasive isolates
of Streptococcus pneumoniae: Alaska, 1991-1998. J Infect Dis 2000;82:490-496.
6. Baggett HC, Hennessy TW, Leman R, et al. An outbreak
of community-onset methicillin-resistant Staphylococcus aureus skin
infections in southwestern Alaska. Infect Control Hosp Epidemiol 2003;24:397-402.
7. Parkinson AJ, Gold BD, Bulkow L, Wainwright RB, et al.
High prevalence of Helicobacter pylori in the Alaska Native population
and association with low serum ferritin levels in young adults. Clin Diagn
Lab Immunol 2000;7(6):885-8.
8. McMahon BJ, Hennessy TW, Bensler JM, et al. The relationship
between prior antimicrobial use, antimicrobial resistance and treatment outcome
for Helicobacter pylori infections. Ann Intern Med 2003;6(139):263-9.