July 23, 1999
Summary
At its meeting in April 1998, the Cell Biology and Biophysics (CBB) Subcommittee of the National Advisory General Medical Sciences (NAGMS) Council identified a need for better methods for examining large macromolecular assemblies and for imaging macromolecules in vivo. Biological structure on these scales is not accessible to analysis by X-ray crystallography or NMR, but is approachable by high-resolution electron microscopy (EM). This workshop was convened to evaluate prospects for utilizing EM to (1) determine the atomic resolution structures of isolated macromolecular assemblies in solution and (2) determine 3-D macromolecular arrangements in cells in conjunction with the known atomic structures of isolated components.
Session 1 examined the hierarchies of biological structure and the capabilities and complementarily of existing methods. Session 2 assessed current capabilities and limitations of EM for structure determination of macromolecules, macromolecular assemblies, and cells. Session 3 defined the research needs of molecular cell biology for structural information and set goals for the determination of atomic structures of isolated assemblies and macromolecular arrangements in cells. Session 4 identified and classified into five general categories the technical problems that must be solved to achieve the goals. Session 5 assessed the technical problems, prospects for solutions, and the types of research effort and resources needed to approach them. Session 6 defined the research approaches needed and the type of support required.
The panel recommended adapting proven EM technology for practical use. Existing methods for intermediate and high-resolution structure analysis of frozen-hydrated specimens should be improved and their throughput increased. The technology of cellular tomography of fixed, stained, sectioned, and embedded specimens should be improved and automated. Improved methods for rapid freezing (to capture transient processes) and for specific labeling and detection of individual proteins in cells should be developed.
The panel concluded that routine atomic structure determination of isolated macromolecular assemblies should be achievable with the next generation of electron microscopes and supporting advances in current technology. The long-term goal of developing knowledge-based EM methods that can define macromolecular 3-D arrangements and interactions in frozen-hydrated cells was considered to be at least partly achievable, but much more challenging. All goals will require major research and development efforts. The biggest challenges will be in automation, detector technology, algorithm development, and high-throughput computation.
The panel recommended support for acquisition of the next generation of electron microscopes and their operation, especially high-caliber personnel. Another priorityis technology development; support for projects targeted to instrumentation and software development is essential. Appropriate peer review arrangements should be made for research and development (R&D) applications.
Background
The NAGMS Council-CBB Subcommittee
At its meeting in April 1998, the CBB Subcommittee of the NAGMS Council noted that integrative approaches to cell function would soon supplant current reductionist and genomic approaches, which are already moving towards completion.
Unfortunately, in many critical emerging areas of integrative cell biology, investigators are working blind, without the ability to directly visualize molecular interactions. The group noted that improved methods for examining large macromolecular assemblies and determining structural arrangements in cells are needed if optimal progress is to be maintained. They singled out EM as a critical technology that requires attention to meet this need.
The CBB Subcommittee noted that progress in some areas is already hindered by lack of tools to define molecular arrangements. Their observations and recommendations were:
(1) Equipment. Upgrading to the next generation of electron microscopes is essential, but these instruments are too expensive ($2-3 million) for existing funding mechanisms. NIH and other agencies should update their equipment funding mechanisms to meet this need.
(2) Personnel. Microscopes should be shared for cost effectiveness and managed for ready access and efficient use. Permanent, highly skilled, career-track support staff is essential to ensure optimal performance and availability. NIGMS' support for synchrotron support personnel was cited as a model for the type of support needed.
Workshop Background
NIGMS staff scheduled this workshop to address the issues raised by the CBB Subcommittee. The primary purpose of the workshop was scientific strategic planning--to identify needs, set goals, identify the problems, and design approaches to solve them.As a starting point to focus discussion, the scientific needs cited by the NAGMSC-CBB panel were formulated by the organizers as short- and long-term goals. At the workshop, participants evaluated these goals. To clarify the organization of this report, the panel's discussions are presented in a framework of four goals:
(1) Automate and increase the throughput of proven methods for intermediate and high-resolution structure analysis of frozen-hydrated arrays and symmetrical particles.
(2) Utilize EM to determine the atomic resolution structures of large, isolated frozen-hydrated macromolecular assemblies in solution, eliminating current dependence upon specimen periodicity.
(3) Extend the imaging capabilities of conventional thin-section EM to 3-D by automating the proven technology of cellular tomography of fixed, stained, sectioned, and embedded specimens. Develop improved methods for (a) rapid freezing (to capture transient processes), (b) uniform preservation and staining of specimens, (c) specific labeling and detection of individual proteins in cells, and (d) display and interpretation of 3-D images.
(4) Determine 3-D macromolecular arrangements in cells by matching electron image data from natively preserved cells with the known atomic structures of isolated components.
Welcome: Wah Chiu and Steve Harrison
Drs. Chiu and Harrison outlined the goals of the workshop and explained the rationale of the agenda in terms of identification of research needs (Session 1), current capabilities and limitations (Session 2), goals (Session 3), technology and technical problems to be solved (Session 4), solutions (Session 5), and plan of execution (Session 6).
Session 1: Research Needs
Moderator: Susan Taylor
Cellular Perspective: Dick McIntosh
In the 1950s, EM defined cell structure at the organelle level and established the foundation for our present understanding of cell function. In recent years, EM has played a reduced and more peripheral role. The reasons for the hiatus were two-fold. First, molecular, cellular, and structural biologists were occupied identifying, dissecting, and characterizing the macromolecules of cells. Second, the limitations of existing EM technology had been reached, and its capabilities to produce fundamental new insights were largely exhausted (a notable exception was the quick-freeze, deep-etch approach developed by Heuser in the 1970s).
Cell biology has now advanced to a stage where information about isolated macromolecules and assemblies must be integrated to define higher-order cellular functions. Optimally, this integrative phase will be spearheaded by structural approaches that can directly visualize cellular organization at resolutions high enough to define macromolecular arrangements and interactions. Alternative (non-structural) approaches are not obvious, but would certainly be indirect, slower, and much more expensive.
EM is the obvious candidate for the structural approach to integrative biology, but significant advances in its capabilities will be required. There is a pressing need for improvements that will make EM good enough and efficient enough to provide the needed information at an appropriate rate.
The first step is to extend the imaging capability of EM from 2-D to 3-D by cellular tomographic methods. Tomography is a high-dose methodology that requires radiation-resistant specimens. To make the most of it, better methods of preparing sections of cells are needed: rapid freezing methods, improved fixation and embedding (especially water-based embedding media), better preservation of structural detail with improved stains, and specific labeling methods for localizing individual molecules in cells.
The ultimate goal is to image natively preserved cells, placing atomic resolution pictures of macromolecules into the cellular images with confidence. Existing conventional high-dose tomographic approaches will not suffice for this goal; new experimental and computational approaches to determining 3-D cellular structure will be required.
Applications of cellular tomography to molecular cell biology were illustrated. These included 3-D reconstructions of kinetochores of PtK cells in metaphase and of part of the Golgi apparatus of NRK cells. The kinetochore map shows several microtubules attached to the outer plate of the kinetochore. These are involved in pulling the chromosome toward the pole of the mitotic spindle. The microtubules are flared outward at their ends in the kinetochore, a morphology characteristic of disassembling microtubule ends. Microtubule dynamics at the kinetochore are the subject of intense investigation in a number of labs. Extensive molecular data are available, and large-scale dynamics can be followed by light microscopy. At intermediate levels of molecular organization--machines and assemblies of machines--the cell biologists must work by touch, rather than by sight. Interactions between macromolecules must be detected indirectly, for example by biochemical or molecular genetic approaches. Being able to actually see the process, for example, by labeling the kinetochore kinesins that have been implicated in microtubule disassembly, would have a major impact on research efforts in the field. Investigators would be able to formulate more specific hypotheses, allowing them to narrow the scope of their inquiries.
The Golgi maps clearly show membranes of individual Golgi and ER cisternae, ribosomes, clathrin and non-clathrin budding profiles, and free vesicles. The membrane dynamics can be observed in unprecedented detail, but much more information is needed. Intensive work over two decades has identified many of the molecular players in the ER and Golgi, but the mechanisms driving the membrane dynamics are not yet understood. As in the case of the kinetochore, investigators are blind at key levels of structural organization and must work by touch, using indirect methods to infer interactions. There is an immediate need for improved structural information to provide a spatial framework for integrating the molecular and functional information. More information about the 3-D arrangement of functional elements is needed to support efficient experimental approaches to the mechanisms of membrane dynamics and molecular flow.
The improvements necessary to visualize, identify, and characterize macromolecular complexes in the context of a cell are easy to define from current knowledge. Significant benefit would result from better methods for tomography with labeled polypeptides and/or oligonucleotides in plastic-embedded specimens; effective techniques for cryotomography of frozen-hydrated specimens at sub-nanometer resolution would really solve the problem. Implementation of these improvements will be a multi-partite challenge, but it is reasonable to expect significant progress on the first part of this effort within 5 years. Bringing cryotomography to the stage where it can provide the resolution we need will take longer and will require improvements in both current and soon-to-be developed technologies, including cryoimaging at very low temperatures, sensitive, low-noise electron image detectors, and methods for image processing.
Molecular Perspective: Steve Harrison
How can the problems of cellular structure and function be defined and dissected? Fundamental cellular processes are not mediated by random interactions of molecules wandering around the cytoplasm, plasma membrane, or organelles. Rather, they are carried out by molecular machines, highly organized yet dynamic macromolecular assemblies, containing numerous protein and RNA subunits. The process of analyzing a molecular machine includes determining the parts list, developing an integrated description of its states, and finally, defining how it works (the mechanism).
In practice, combination of complementary EM and X-ray/NMR analyses is an efficient approach, and it is required for the larger machines. In outline, begin with EM to define the framework, use X-rays and NMR for atomic resolution analysis of subassemblies, and then synthesize EM and atomic resolution data to derive the entire structure. For definition of states and dynamics, return to EM to freeze steps in the process and capture short-lived intermediate states. After EM defines the large-scale structural transitions, return to atomic resolution methods to define the underlying molecular transitions and complete the picture. To fully define the structural basis of function requires not only the three dimensions of space, but the fourth dimension of time. Time-resolved molecular movies must define meaningful intermediate states and the structural transitions between them.
Structural biology has now advanced to the solution of small machines--topoisomerase, GroEL, F1 ATPase, ribosomes, and coated vesicles. Next on the agenda are larger machines, including the control boxes or logical machines involved in transcriptional control ("enhanceosomes") and signaling and regulation ("signalsomes"). One example is the T-cell signaling complex, where the goal will be to discover the combinatorial logic underlying its functional behavior. In approaching these problems, EM will play a role in defining the direction of scientific inquiry--it is important to be able to see structure in order to determine how to proceed.
Features of the complementary structural approach were illustrated for Gro-EL, the nucleosome, topoisomerase II, rheovirus, and the clathrin coat. Technical issues addressed included: bootstrapping of X-ray phases, single particle analysis, symmetric and asymmetric structures, throughput (including automation of image analysis), and the superpositioning of EM and X-ray data. Issues in the logic and strategy of experimental design included: the role of EM in identifying problems and providing a framework for experimental design, iterative approaches to dynamics and mechanism, and the roles of mass spectroscopy and recombinant technologies for expression and reconstitution of parts.
Session 2: Electron Microscopy Capabilities and Limitations
Moderator: David DeRosier
This session provides the technical background for the subsequent strategic planning sessions. This synopsis of the presentations focuses on the EM related aspects. For biological background, significance, and conclusions see the abstracts in the Appendix and the literature references therein.
Instrumentation
Electron Microscope
Electron Source
Types are conventional (filament, LaB6) and FEG (Field Emission Gun).
The key property is coherence of emission, critical for high resolution.
Specimen Support Film
A thin carbon film made by vacuum evaporation, usually holey, which holds the specimen.
Specimen Grid
A disposable fine copper screen that holds the support film.
Specimen Holder
Cryo or conventional, holds the grid, is inserted into the stage.
Specimen Stage
Part of the microscope column, located between the condenser and objective lenses. Stages may be either conventional or cold. Cold stages are cooled by either liquid nitrogen or liquid helium. Stages may also be either fixed-angle or tilting. Tilting stage types include single axis, dual axis. Tilt capability ranges up to +/-70°.
Electron Lenses
The critical lens is the objective, which forms the image. A key property is correction of aberrations (spherical, chromatic etc.).
Electron Detector
Photographic film or electronic (CCD or other technology). Key properties are: (1) MTF (Modulation Transfer Function), fraction of signal detected as a function of spatial frequency (resolution) at the image plane, (2) size (of pixel array), (3) noise, and (4) readout time.
Other Apparatus
Cryo Freezing and Transfer Apparatus
For the freezing and transfer of specimens.
Ultramicrotome
Cuts sections from plastic embedded or frozen sections.
Film Scanner (Densitometer)
Reads optical density of processed film.
Radiation Damage
Radiation damage ultimately limits what can be accomplished with EM. Most electrons pass through the specimen without imparting any energy to it, but a few collide inelastically. These collisions transfer energy to the specimen, disrupting its chemical structure. After a relatively short exposure to the beam, an individual macromolecule is destroyed by inelastic collisions. This occurs long before enough electrons have passed through to allow determination of its structure ab initio (i.e., from scratch, unassisted by other sources of information). There are three ways to detour around the problem of radiation damage:
(1) Chemically convert the specimen to a radiation resistant form, usually by fixing, staining, plastic embedding, thin-sectioning, and post-staining. For conventional thin-sections, the harshness of these procedures limits the resolution of reproducible detail to 30-60 Å. We refer to this approach as conventional thin-section EM.
(2) Expose many identical molecules for only a short period of time, collect partial data from each of them, and computationally combine the data. From the combined data, calculate the average structure of the set of molecules. Computational averaging is the approach underlying all current high-resolution work. This is a proven approach for atomic resolution.
(3) An alternative approach is to expose a single specimen for a short period, and then match the resulting incomplete electron image data with information derived from other sources to identify the structure. Thousands-fold fewer electron data are required to recognize a molecule than to determine its structure from scratch. This is the rationale behind Goal 4, determining macromolecular arrangements in native cells. Knowledge-based analysis is an unproven approach, but may be the only way to derive atomic resolution structure from unique specimens by EM.
Image Processing
Basic image processing for EM involves two major classes of operation, 3-D reconstruction and computational averaging.
3-D reconstruction is the process of deriving a 3-D density map from a series of views of the specimen seen from different directions. Depending on specimen geometry, tilting of the specimen grid in the microscope may or may not be required to record the needed range of views.
Computational averaging is the process of imaging many molecules that have identical structures, and then averaging them. This works around the problem of radiation damage. Averaging more molecules improves the signal-to-noise ratio and extends the attainable resolution.
The resolution attainable for a specimen by computational averaging depends upon the number of identical molecules with images that can be recorded and computationally averaged together. The numbers below are what are typically attained in recent practice. Improvement of technology to reduce signal losses would lower the number of molecules needed.
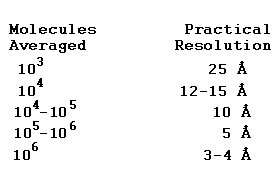
Specimen Symmetry and Redundancy
The six presentations will illustrate the major classes of specimen symmetry and redundancy. They begin with the most structurally redundant specimens, 2-D crystals, proceed through specimens with lower levels of redundancy, and finish with a cellular organelle, which has zero redundancy (every specimen is structurally unique). Note the striking differences in specimen handling, data collection, and image processing. Note that all specimens, except the last (the centrosome), were quick-frozen in the absence of stain and are in a native (frozen-hydrated) structural state.
Defined and Variable Structures
Structures under 107 daltons (e.g., virus particles, ribosomes) are often well defined. Larger assemblies (for example, actin bundles) are more variable in size and structure and may incorporate a polymorphic mixture of symmetries. They are not tractable to high-resolution analysis by existing methods and will require fundamentally different approaches.
Classes of Defined Structure: Symmetric and Asymmetric
Symmetric structures have regular geometric repeats. Asymmetric structures have no regular repeats. The ribosome is an asymmetric structure.
Symmetric Structures
Symmetric structures may have (a) point, (b) 1-D, (c) 2-D, or (d) 3-D symmetries.Viruses and many protein complexes have point symmetry (e.g., hepatitis virus, icosahedral, ryanodine receptor, 4-fold rotational). Actin filaments and microtubules have 1-D symmetry (a regular helical geometrical repeat). Crystalline tubulin sheets have 2-D symmetry.
Tubulin: Ken Downing
Tubulin's strong tendency to polymerize has made it so far impossible to obtain crystals for X-ray diffraction and also provide a type of crystalline sheet that is ideally suited to study by electron crystallography. The tubulin sheet is a 2-D crystal, one molecule thick, and typically 1 to 2 microns in extent in the other two dimensions. The microscopic crystals are prepared from depolymerized tubulin in the presence of zinc and taxol. They were used to solve the tubulin structure to a resolution of 3.7 Å.
Crystals in suspension were quick-frozen, and the specimen grids were inserted into a tilting cryostage. The microscope, a JEOL-400 with a liquid nitrogen cooled cryostage and a LaB6 electron source, was operated at 400kV. About 100 images, each with about 10,000 molecules, were recorded on film at different tilts up to 60°. Only the first image recorded from a specimen was used for data reduction.
Each image produced one average projected (2-D) view through the structure, from which crystallographic phases were derived. Diffraction intensity data were collected from the same type of 2-D crystals by operating the microscope in diffraction mode. Although the intensities could also theoretically be derived from the images, separate collection by diffraction avoids complicated corrections and is more accurate and efficient. The diffraction data and 100 projected views were combined to produce the 3-D density map. The number of individual molecular images computationally combined to derive a structure exceeds 1,000,000.
The resolution of the map was 3.7 Å, but it was of much higher quality than an X-ray map based on data from crystals that diffract measurably to only 3.7 Å. The high quality results from extremely accurate determination of the crystallographic phases. An atomic model was built into the density map using conventional X-ray model-building software. Because the phases were so accurate, phase refinement was not required to obtain an interpretable map.
This crystalline form of tubulin was discovered over twenty years ago, but only recently has the methodology of electron crystallography been developed to the point that it appeared worthwhile pursuing the structure. The data collection and associated development work took 5 years, but the data collection phase could now be repeated in one year. As the techniques and instrumentation continue to develop, we will be able to solve structures of similar crystalline specimens, such as integral membrane receptors and signaling molecules, far more rapidly and to higher resolution. Electron crystallography is already a viable complement to X-ray crystallography for determining structures of proteins that are refractory to conventional crystallization.
Problems that spoiled images included faults in the crystal lattice, imperfect flatness, beam-induced effects, stage drift, and imperfect focus. Image processing proceeded at the rate of one image or ten diffraction patterns per day, and it would be a bottleneck if the analysis were repeated.
The measured data from images were much weaker than their theoretical value, with over 90 percent of the signal being lost to various causes. Some of the problems probably originate at the specimen and stage, perhaps with stage drift or beam-induced effects. These issues were revisited in detail in Session 5-2.
Acetylcholine Receptor: Nigel Unwin
Acetylcholine receptor membrane tubes are prepared from eel electric organ membranes. They are 2-D crystals rolled into tubes. The typical extent along the long axis is several microns, and the typical diameter is about 200 nm. For purposes of data collection and analysis, the specimen is a helix rather than a crystal. Because of the helical arrangement, a complete set of views of the receptor covering 360° of tilt is seen in a single image of a tube.
The membrane suspension on a grid is dropped into liquid ethane at -160°C. To capture short-lived conformational states, the grid is sprayed with acetylcholine as it drops towards the dewar. By changing the height at which the grid is sprayed, the time that the receptor is exposed to ligand before it is frozen is precisely controlled. The structure can be frozen in the different conformational states corresponding to short-lived events in ligand binding, channel opening, and desensitization.
The microscope was unique in three respects: (1) a liquid helium (4°K) cooled specimen stage, (2) 300kV operating voltage, and (3) a field emission tip in the electron source. The instrument was constructed by Dr. Fujiyoshi and his father, a retired machinist, in collaboration with the JEOL Company of Japan. The cryochamber was machined from a single billet of copper alloy by Fujiyoshi Sr., eliminating the need for soldered joints. Leaky joints had defeated previous attempts to construct a practical helium-cooled electron microscope stage.
All of the data were collected in Japan. Over a period of 3 years, Dr. Unwin made visits from his home laboratory in England to Fujiyoshi's lab in Japan every 6 weeks on average, with a typical stay-over of 7 to 10 days per visit. He has recently purchased an apartment near the lab. Dr. Unwin said that this work would have been impossible on any other microscope. Critical attributes of the instrument include specimen cooling, high beam coherence, and optimal accelerating voltage. He noted that there are no microscopes like this one in the United States. At this time, there are several in Japan, and they are now available commercially.
Images (on film) of about 500,000 molecules were combined and thousands of layer lines collected to produce a 4.6 Å resolution 3-D density map. Density features corresponding to elements of secondary structure can be observed, but the resolution is not yet high enough to allow an atomic model to be built. Conformational changes to the ion channel resulting from ligand binding are clearly observed. An atomic structure is anticipated in the next few years.
Dr. Unwin noted that the measured data was at about 20 percent of its theoretical signal strength. Pressed to speculate on signal loss, he tentatively attributed fractional losses to six factors, none greater than 30 percent. These losses were: (a) the modulation transfer function (MTF) of the film and densitometer (30 percent loss); (b) partial coherence of the electron source (30 percent); (c) uncorrected disorder in the specimen lattice (30 percent); (d) depth of focus (the upper and lower sides of the specimen tube are 100 nm apart along the microscope axis) (20 percent); (e) beam tilt (20 percent); and (f) radiation damage (10 percent). He thought he had controlled the effects of specimen charging, and he observed no specimen movement. He attributed no signal loss to these causes.
Thousands of images were recorded. Almost all had to be discarded because of flaws in the crystalline lattice or other specimen defects. Dr. Unwin noted that the dependence of data collection on the near perfect preservation of periodic structure of an extremely delicate specimen was the Achilles heel of the project.
Dr. Unwin made two points in closing his scientific presentation: (1) it is not possible to do this kind of analysis without truly top class instrumentation and (2) it is not easy to explore structural transitions of membrane proteins in such detail by other methods.
Next-Generation Microscopes
Depending on the experimental parameters like specimen thickness, some projects can be taken to atomic or intermediate resolutions on microscopes currently available in the United States. In these cases, collecting more data can partially compensate for deficiencies in performance. However, it may be necessary to collect and process two to three times more images, increasing the duration and cost of the analyses several-fold. At this time, the cost of the next generation of 300kV/FET/liqHe microscopes is too high for American investigators. Adjustments are needed so that U.S. investigators can have access to these instruments.
Technology Development: United States and Abroad
Dr. Unwin, an NIGMS grantee, is now at MRC Cambridge, England, but was formerly a professor at Stanford. He was asked to compare and contrast the state of high-resolution EM in Japan, Europe, and the United States. He noted that the U.S. system has fostered the growth of a strong community of independent investigators and provides the best opportunities for new PIs. The new PIs are its key strength. However, the U.S. system of short-term support does not specifically provide for long-term work.
The European and Japanese systems make specific accommodations for support of long-term technical development. Financial support for technology development is particularly strong in Japan. In Europe, centers like the Max Planck also provide long-term support for technology R&D. Participants noted that the short-term funding cycle in the United States often does not reward investment in long-term goals.
Hepatitis B Virus Capsid: Alasdair Steven
The hepatitis virus coat is a symmetric particle comprising 60 equivalent asymmetric units, arranged in icosahedral symmetry. There is structural redundancy within each particle and also between particles. Virus particles in frozen-suspension on a holey carbon film were inserted into the liquid nitrogen-cooled cryostage and imaged on film in a Philips CM20-FEG at 120kV. Careful correction for the Contrast Transfer Function (CTF) was necessary for each image. Because the virus particles are in random orientations, a complete set of views from all orientations is available without the use of a tilt stage or holder.
The number of particles computationally combined was over 2,000; after application of the symmetry, these yielded over 105 molecular images. The process of particle selection was partially automated. Because of the symmetry averaging, only the icoashedrally repeating structural features of the virus coat are seen. The averaging operation blurs features in the asymmetric interior, so no detail is seen there. The resolution, about 8 Å, is high enough to identify major elements of secondary structure, like extended alpha-helices. In this map, a four-helix bundle, known from an X-ray structure, can be identified with a feature in the EM density map.
The analysis was assisted by labeling combined with difference imaging. The C-terminus was located by engineering in a cysteine and labeling it with undecagold, an 11-atom gold cluster. A hexapeptide in the sequence was labeled with Fab fragments of a specific monoclonal antibody. The N-terminus was visualized by appending an octapeptide, which was visible in a difference map.
The resolution of virus structures improved from 30-40 Å in the early1980s to 15-10 Å in the early 1990s and is still rapidly improving. In 1997, three structures at 7-9 Å resolution were published. Accurate 3-4 Å resolution determinations may be sufficient to define atomic arrangements. This goal is certainly attainable, perhaps relatively soon.
Scientific horizons include: (a) analysis of large, rare, and experimentally wayward viruses, (b) analysis of drug binding for vaccine development, (c) analysis of precursors and structural transitions in virus assembly and maturation, (d) imaging of the asymmetric components of symmetric viruses, and (e) analysis of fully asymmetric viruses (e.g., Ebola, hemorragic fever viruses).
Dr. Steven noted that the measured data was at about 10 percent of its theoretical signal strength. Improvement of the technology to address the causes of degraded performance should raise this value to over 50 percent. To accomplish this, the needs are: (a) better cold stages (using liquid helium), (b) more coherent electron sources (improved FEGs, field emission guns), (c) better electron image recorders (using CCDs, charge-coupled devices, rather than photographic film as the electron detector), (d) improved corrections for geometric distortions and temperature factor, and (e) automated data collection, particle selection (detection and rejection of imperfect particles), and data reduction.
Ribosome: Joachim Frank
The ribosome is the molecular machine par excellence. It is an asymmetric particle, but its structure is well defined so that data from different particles can be combined. Isolated ribosomes complexed with combinations of fMet-tRNA, elongation factor G, and mRNA in frozen suspension were inserted into a liquid nitrogen-cooled cryostage. Images (on film) of between 10,000 and 15,000 particles were computationally combined. The electron microscope and film scanner used for the work are both about 10 years old. Because the particles are in random orientations, a complete set of views of the particle covering all orientations is available without the use of a tilt stage or holder.
Since the particles were all in different random orientations, the orientation of each particle had to be determined in concert with all the others by an iterative classification and refinement procedure. This approach of single particle 3-D reconstruction has been developed in the Frank lab over a period of 20 years, in parallel with the ribosome project. Resolution is evaluated by dividing the data into two independent data sets and following their correlation as a function of resolution.
There are now 3-D density maps of the ribosome in various binding states at resolutions ranging from 15 to 20 Å. These provide a picture of the complete elongation cycle. The fMet-tRNA, elongation factor G, and mRNA have all been fitted into the structure. The cryo-map has also been used in the phasing of X-ray data, resulting in a 9 Å map of the large ribosomal subunit recently published in collaboration with the Moore and Steitz labs at Yale.
The current 15 Å resolution of the analysis should soon be extended to about 12 Å (note, in July 1999, the resolution had reached 11.5Å). The next resolution barrier will be conformational heterogeneity of the particles. This problem will be addressed by conformational classification methods. There are many conformations, and not all of them are meaningful. It will be necessary to identify the functionally significant structural states. Much work is necessary to approach the resolution range (5-8 Å) that allows elements of secondary structure to be identified, and even more work is needed to approach atomic resolution. However, even the present medium resolution (15 Å) density maps allow atomic structures to be derived by combining complementary EM and X-ray data.
Challenges for the future include mapping of initiation, release and recycling factors, and time-resolved imaging of structural events in the elongation cycle. The end product will be a 3-D molecular movie on a millisecond timescale. A technical hurdle to overcome either experimentally or computationally will be occupancy of states. Either specimen preparations yielding high occupancy of specific states, or an increased investment in data collection and classification algorithms will be needed. What will it take? Certainly, staff will be needed who can handle this type of challenge. How will they be trained? How can needed collaborators be recruited?
Technical problems on the specimen and data collection side include: (a) lack of reproducibility in specimen preparation, especially ice thickness, (b) performance of the relatively old microscope, and (c) lack of automation in data collection (operating the controls and the loading, processing and scanning of photographic films). Problems with image processing and data reduction include: (a) lack of rigorous methods for classifying particle heterogeneity, (b) difficulty of handling large data sets (database management tools are lacking), (c) clumsy, outdated, inefficient user interfaces in the software, and (d) lack of sophisticated modeling tools (estimated cost to develop $10 million). The underlying cause is shortness of staff in the area of algorithm development and scientific programming. Personnel designated for these tasks are typically deleted from budgets by study sections.
Cytoskeletal Complexes: Ron Milligan
Actin and tubulin are helices, 1-D crystals of indefinite extent along the long axis. Complexes of actin filaments and microtubules with the motor proteins myosin and kinesin are prepared from purified components under defined conditions of nucleotide concentration and ionic strength. Complexes in frozen-suspension on a holey carbon film are inserted into the liquid nitrogen-cooled cryostage and imaged on film in a Philips CM20-FEG. Undistorted stretches of motor decorated filaments are selected, imaged on film, and processed. Because of the helical topology, a complete set of views of the motor filament complex covering 360° of tilt is available for 3-D reconstruction in a single image of a decorated filament, without the use of a tilt stage or holder. The helical symmetry is also exploited for computational averaging. The number of molecular images computationally combined is 1,000 to 3,000.
Helical image analysis produces 3-D density maps of the actin-myosin and microtubule kinesin complexes at 20-30 Å resolution. Atomic resolution structures of actin, myosin, tubulin, and kinesin are available from X-ray and electron crystallography. The atomic structures are docked into the 3-D maps to provide atomic models of the complexes. By varying the state of bound nucleotide, conditions corresponding to different stages of the force and motion producing cycle have been visualized.
Long-term goals are time-resolved molecular movies of the entire power cycle in both systems, providing a complete mechanistic understanding of mechanotransduction, including the structural determinants of motor parameters like directionality and stroke length. A resolution of better than 10 Å would detect major features of secondary structure and eliminate all ambiguities in the docking of atomic structures into the 3-D EM density maps. For this, improved methods of correcting for specimen disorder computationally are needed.
The microscopy and image processing are labor intensive, slow, and tedious. More automation is needed so that investigators can answer biological questions fast. Currently, an analysis of a single structural state, involving collection and processing of 50 data sets, takes months. The dream is to achieve the same result overnight. Keys will be automation of specimen preparation, microscope operation, and image processing.
Centrosome: David Agard
Centrosomes are not only asymmetric, but each one is also unique in structure. Although all are assembled to the same set of specifications, their construction varies in detail (they are "polymorphic goobers"). All the data needed for 3-D reconstruction must be collected from a single specimen. This rules out the use of unstained cryo-specimens, which are too radiation sensitive to survive the number of exposures required. Whole cells are fixed, stained, and plastic embedded in the traditional fashion, and thin-sectioned on an ultramicrotome in the usual manner. Native fine structure at high resolution is not preserved, but the purpose of the analysis is to define the large scale architecture of the organelle.
The specimen grid holding the section is clamped in an adjustable tilt holder in a conventional microscope tilting stage (not a cold stage). A region of the section that passes through the organelle at the desired level is selected. A large number of images (>120) is recorded over an angular range of +/-70°. These are combined by 3-D reconstruction to generate a 3-D density map of the stain distribution in the section though the organelle. There is no computational averaging. This type of analysis is accurately described by the term cellular tomography.
The resulting 3-D map is messy and difficult to interpret, due to uneven specimen staining and preservation. Nevertheless, it reveals structural detail unattainable by any other approach. Despite its comparatively low resolution (limited by the 30-60 Å reproducibility of specimen preservation), the 3-D map provides a structural framework that can address important practical questions (in this case, about microtubule nucleation and centrosome function in vivo). Even large organelles can be tackled in segments by a "divide and conquer" approach. In combination with specific labeling, cellular tomography is an extremely powerful structural approach.
Cellular tomography extends the imaging capability of conventional EM, allowing it to resolve detail in the third dimension (along the optical axis). Comparison of cellular tomography with confocal microscopy is apt. Wider application of cellular tomography, at its current level of development, could have a significant impact in a number of areas of molecular cell biology. However, a large staff and further development of supporting automation would be required to run a center specializing in cellular tomography.
Data are still collected on both film and electronically by CCD. Film can record much larger areas, but the labor overhead is huge. The CCD eliminates handling and scanning of films, but has a small active area (1K x 1K pixels) and can only record a small area of the specimen at a time. The control of the microscope during the process of data collection has been extensively automated. Despite progress on automation, the repetitive work is still very labor intensive and boring.
To bring the capabilities of cellular tomography into the research mainstream requires major upgrades and development. These include: (a) higher accelerating voltage, allowing thicker specimens to be imaged, (b) two-axis, high-tilt specimen holders, and (c) CCD detectors with larger active areas. For data collection, methodological improvements are needed in (a) automation of microscope operations and (b) dynamic drift correction. Improved alignment of images via improved iterative refinement methods is needed for 3-D reconstruction. For the understanding and communication of results, better methods must be developed for assisting interpretation of 3-D density maps, including ways of objectively converting them into comprehensible display formats.
The next stage of development must also concentrate on improving specimens: (a) improvement to traditional fixing, staining, and embedding methods to improve uniformity of preservation and staining, (b) specific labeling methods so individual proteins can be located, and (c) rapid freezing methods.
Session 3: Goals
Moderator: Paul Matsudaira
Molecular cell biology and its ability to order the information revealed by genomics, genetics, and structural biology is steadily moving into a new era of integrative cell biology. Three classes of problems are emerging as particularly worthy of attention: (1) a need to map atomic structures onto high-resolution models of non-crystalline cellular machines and assemblies, (2) an anticipated requirement to relate light microscopy of dynamic cell processes with molecular level information, and (3) a capability to relate genomic information to cellular structures. Each of these issues can be addressed by use of modern techniques of EM. However, an enormous gap is present in our ability to merge atomic level information from X-ray crystallography and NMR spectroscopy with genetic, biochemical, and light microscopic information. The electron microscope can bridge this gap in information if a few technical advances are made. Without this tool, molecular cell biology will be unable to move into the next dimension, an integrative examination of cell structure and function, and functional genomics will not be able to parlay the wealth of genomic information to cell function.
The purpose of Session 3 is to identify research needs in biology that require EM and set goals for developing the capabilities needed. To accomplish this, this session first addresses four representative integrative problems in molecular cell biology and the capabilities and limitations of the technologies that EM must interface with.
The presentations examine how complementary technologies are applied in integrative cell biology. All four are problems of higher order cellular function: motility, organelle dynamics, membrane transport, and regulation. Conceptually, all four problems require resolution of hierarchical levels of structural organization and function. Capabilities and limitations in the methods available for covering the different hierarchical levels are highlighted.
The presentations address all levels of structure and function: (1) individual component proteins (functional and structural domains of protein kinase A and neonatal Fc receptor, FcRn), (2) protein-protein interactions (receptor dimerization, receptor-ligand binding, kinase-anchor binding), (3) macromolecular assemblies (oligomeric ribbons, dynein arms, nuclear pores), (4) organellar sub-domains (axonemes and basal bodies of flagella, membrane domains of the ER and Golgi), (5) whole organelles (ER, nuclear envelope), and (6) whole cells (disassembly and reassembly of membrane systems in mitosis).
Motility: Mary Porter
The flagellar axoneme is a highly organized motile organelle exhibiting an unusual degree of structural redundancy. It is a cylinder about 200 nm in diameter and about 10 microns in length. There is an axial structural repeat every 96 nm and an approximate 9-fold radial symmetry of the outer arms. The most abundant axonemal components are tubulin and dynein, which are responsible for its core activity. The axoneme contains at least eight different dynein complexes, and each one is a large assembly (2 x 106 daltons) composed of 3-15 polypeptide subunits, the largest of which is the heavy chain (about 5 x 105 daltons). Other axonemal proteins regulate motility or mediate assembly and maintenance of the organelle.
Investigators initially approached the axoneme from the starting point of whole cell function and genetics. Genetic analysis of assembly and motility defective mutants has led to the cloning of more than 45 genes encoding flagellar components. Phenotypic analysis has also identified the approximate structural location of about 115 out of the estimated 250 different polypeptides that make up the organelle. Light microscopic and biophysical studies have extensively characterized its very complex repertoire of functional behaviors in vivo. The structure has also been investigated from the molecular direction, beginning with the isolation of dynein by Gibbons in the 1970s. Recently, atomic structures of tubulin and some dynein light chains have been determined. Others can be approximated by homology modeling, since many flagellar proteins have cellular homologs whose structures are known. The full dynein complex is a likely candidate for atomic structure determination in the relatively near future, probably by X-ray crystallography, assisted perhaps by EM. In the foreseeable future, atomic structures of most flagellar component proteins will be available.
Dr. Porter's group, in collaboration with colleagues at the Boulder HVEM Center, are combining genetics and EM to locate proteins in the structure. Wild type and mutant flagella are conventionally fixed, stained and embedded, thin-sectioned, and imaged in the electron microscope. Non-uniformity of specimen preservation and staining in conventional thin-sections limits the effective resolution to 30-60 Å. The approximate structural repeat is exploited by computational averaging to improve the signal to noise ratio. Average images of wild type and mutant flagella are subtracted to produce difference maps. This approach has located six of the eight dynein complexes in the structure, as well as the location of other regulatory proteins in the dynein arm region.
Full understanding of the function of this organelle, will require definition of the atomic structures of the whole flagellum frozen at the key stages of its functional cycle. It will be necessary to place atomic models of the component proteins into these structures accurately enough to define their molecular interactions. This will probably be accomplished in stages, starting with structures of subassemblies like inner and outer dynein arms determined by complementary X-ray/NMR/EM approaches. Atomic structures of subassemblies will then have to be combined with high-resolution EM data from the intact frozen-hydrated structure, as described for Goal 4. It is likely that the technology to achieve a Goal 4 style synthesis for flagella will lag behind the availability of structures of most of the flagellar component proteins, perhaps for an extended time.
Membrane Transport: Pamela Bjorkman
The neonatal Fc receptor (FcRn) transports maternal immunoglobulin G (IgG) to the bloodstream of the fetus during the acquisition of passive immunity. FcRn also binds IgG and returns it to the bloodstream, thus protecting it from a default degradative pathway. FcRn is structurally similar to class I major histocompatibility complex (MHC) molecules, despite differences in the ligands they bind (the Fc portion of IgG and antigenic peptides, respectively). The X-ray crystal structure of the complex between FcRn and Fc localizes the binding site for Fc to the side of FcRn. A dimer of FcRn heterodimers observed in the co-crystals and in the crystals of FcRn alone could be involved in binding Fc, correlating with the 2:1 binding stoichiometry between FcRn and IgG and suggesting an unusual orientation of FcRn on the membrane.
Careful biochemical and biophysical and structural investigations of native and mutant receptor function have accumulated a wealth of circumstantial evidence suggesting that oligomerization of FcRn and IgG into zig-zag ribbons parallel to the membrane surface may be an important step in transcytosis. Reconstitution in vitro generates ribbons that are clearly visible by EM, but electron microscopic approaches for examining the corresponding process in vivo are lacking.
Organelle Dynamics: Jennifer Lippincott-Schwartz
The major membrane systems of eucaryotic cells comprise a dynamic network with bidirectional membrane transport pathways and overlapping compartmental boundaries. Membrane traffic and organelle biogenesis/maintenance are fundamentally linked within this system, with perturbations in membrane traffic quickly leading to changes in organelle structure and identity. Dissection of the molecular basis of these properties in yeast and mammalian cells is a central problem of integrative biology.
An important new tool for investigating how the organelles receive cargo and maintain their integrity in the face of ongoing traffic flows has emerged with the advent of green fluorescent protein (GFP) chimeras. GFP chimeras, which can be visualized in the unperturbed environment of a living cell, are being used in a wide variety of applications to study organelle dynamics. These include time-lapse imaging, double-label, and photobleach experiments. These studies are helping to clarify the steps involved in the formation, translocation, and fate of transport intermediates associated with organelle compartments, including the roles of cytoskeletal elements. They are also providing insights into mechanisms of protein retention and localization within membranes.
Quantitative time-lapse imaging data of single cells expressing the transmembrane protein, vesicular stomatitis virus ts045 G protein fused to green fluorescent protein (VSVG-GFP), were used for kinetic modeling of protein traffic through the various compartments of the secretory pathway. A series of first order rate laws was sufficient to accurately describe VSVG-GFP transport, and provided compartment residence times and rate constants for transport into and out of the Golgi complex and delivery to the plasma membrane.
GFP variants with shifted spectral characteristics offer enormous potential for double-labeling experiments and protein-protein interaction studies. Photostable spectral GFP variants (W7 and 10C) have been used in dual-color, time-lapse imaging of fusion proteins in living cells by both wide-field and confocal microscopy. They were highly photostable during repetitive long-term imaging and are cleanly separated by their different excitation spectra alone with negligible crossover of fluorescence. Time-lapse image sequences of cells co-expressing Golgi and nuclear membrane markers of the Golgi complex (galactosyl transferase) fused to W7 and a marker of the nuclear envelope (lamin-B receptor) fused to 10C provided the first simultaneous visualization of Golgi and nuclear envelope membranes in living cells.
Dr. McIntosh has discussed the role of complementary EM approaches in analyzing underlying mechanisms of ER and Golgi dynamics in Session 1. With the anticipated arrival of dual labeling technology, under development in the Tsien and other laboratories, LM and EM cellular tomography will be able to interface and cover most levels of structural organization in these organellar systems.
Signaling and Regulation: Susan Taylor
Cellular regulation is mediated by complex networks of interacting proteins that translate and integrate signals received at the plasma membrane and deliver them to various sites within the cell. Organizing the locations and regulating the activities of protein kinases is a central function of these networks. Protein kinases comprise a large part of the cell's basic machinery. Two percent of the genome encodes protein kinases (about 2000 of them). They all have similar folds. Protein kinase A (PKA) is one of the simplest and best understood protein kinases. Substantial progress has been made in understanding how it is regulated and localized at the level of the protein itself. Its structure has been determined by X-ray crystallography, and the functions of its subunits and functional domains have been characterized.
Attention is now shifting to higher orders of structure and function. The discovery of A kinase anchoring proteins (AKAPs) revealed a family of proteins that target PKA to various sites within the cell. Two families of AKAPs bind both the RI and RII subunits of PKA. D-AKAP1 targets PKA to mitochondria or ER while D-AKAP2 has an RGS domain. The structures and functions of these are now being determined, as well as the functional consequences on PKA of anchoring reaction.
At the next level of investigation, GFP chimeras are being constructed in pairs to identify physiological binding partners by confocal LM and EM. Brown adipose tissue, where PKA and D-AKAP1 are both expressed at high levels and co-localize to the outer mitochondrial membrane, is being developed as a cellular model. There is an immediate need for the application of cellular tomographic EM methods to correlate membrane dynamics with PKA and AKAP functions.
It is already apparent that the molecular architecture and the combinatorial logic of cellular regulation and signaling are inseparable. Systems that receive, integrate, analyze, and transmit signals perform these functions by delivering receptors, kinases and other signaling enzymes, and regulatory components to specifiable locations. The ability to define the 3-D locations of signaling components dynamically will be central to unraveling the logic of biological computing networks.
Analysis: Paul Matsudaira
The contrast between levels of research activity in EM and the high-impact mainstream technologies of LM and X-ray/NMR is striking. State of the art LM, X-ray, and NMR are employed by hundreds of laboratories, are readily accessible technologies, and are exploited effectively. EM is still an arcane specialty. It's proven capabilities are enormous, but have not been fully exploited in practice. The skill, labor, and expense of state of the art EM puts it out of reach of the scientific mainstream.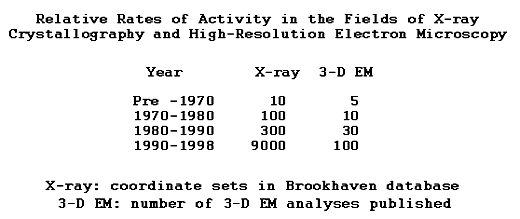
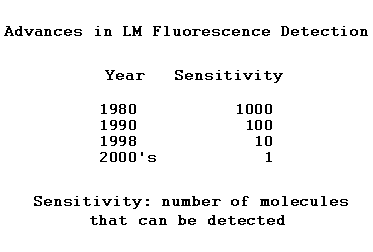
LM technology has been advancing at an astonishing pace since the 1970s. There have been a number of groundbreaking advances in areas of instrument design, labeling, and image analysis, as well as continuous refinements and improvement. These move to market rapidly and see almost immediate practical application in the field as routine tools. Advances in experimental design to exploit the new technology are just as rapid. Technological and conceptual advances are reflected in fundamental new insights and discoveries on an almost yearly basis.
Goals
Short-term goals are to make proven EM technologies into practical tools by improving instrumentation and automation. High-resolution methods have already had a significant impact, but must be developed as routine tools. There is an immediate need for mainstream application of cellular tomography. Long-term goals are to develop the new capabilities that must be achieved to realize the full potential of EM in biology.
Goals for Macromolecules and Assemblies
(1) Frozen-hydrated arrays and particles at high resolution. The technology, presented in Session 2, is proven. Automation and instrument improvements require extensive but feasible research and development to make them into practical tools.
(2) Atomic resolution structures of macromolecular assemblies in solution. Atomic structure determination of asymmetric particles of defined structure in solution by unassisted EM and computational averaging. Elimination of dependence upon specimen periodicity and symmetry to achieve atomic resolution. Term is intermediate, 5 to 10 years.
Goals for Cells
(1) Conventionally embedded and sectioned cells in 3-D at low resolution using cellular tomography (extension of traditional EM to 3-D). The technology, presented in Sessions 1 and 2, is proven. Extensive improvements to automation, instrumentation, and specimen preparation are needed to fully exploit it as a practical routine tool.
(2) Macromolecular arrangements in cells in 3-D by EM and knowledge-based approaches (matching of EM data by pattern recognition to databases of atomic structures of cellular proteins and macromolecular assemblies). Term is long, 10 to 20 years.
Goals for Macromolecules and Assemblies
(1) Frozen-hydrated arrays and particles. Develop high-throughput capability for the proven methods described in Session 2: (a) atomic resolution of 2-D crystals by unassisted EM (e.g., tubulin, anticipated for acetylcholine receptor); (b) composite approximate atomic resolution structures of periodic specimens derived by intermediate resolution EM in conjunction with atomic resolution X-ray/NMR (e.g., f-actin-myosin, microtubule-kinesin); (c) direct atomic resolution of some smaller symmetrical particles (some viruses) by unassisted EM (e.g., anticipated for hepatitis virus); and (d) indirect composite atomic resolution structures of asymmetric particles of defined structure by complementary EM/X-ray/NMR (e.g., ribosome).
(2) Atomic resolution structures of macromolecular assemblies in solution. Atomic structure determination of asymmetric particles of defined structure in solution by EM and computational averaging. Elimination of dependence upon specimen periodicity and symmetry to achieve atomic resolution.
Rationale: At its current state of development, EM is already a useful method for determining the structures of isolated macromolecular assemblies at intermediate resolution. The recent elucidation of the atomic structure of tubulin by EM has highlighted the high-resolution imaging capabilities of the electron microscope and its potential for producing more detailed and accurate structures of large complexes. As currently practiced, EM at resolutions sufficient to define an atomic model (3.5 Å or better) requires an ordered 2-D array analogous to the 3-D crystal required for X-ray crystallography. However, the potential exists to bypass this limitation and collect the required data from randomly oriented molecules in frozen solution, making any isolated macromolecular assembly with a well-defined structure accessible to high-resolution analysis. The goal is to utilize EM to determine atomic resolution structures of macromolecular assemblies in solution.
Outline: For high-throughput analysis of macromolecules and assemblies it is critical to eliminate dependence upon 2-D crystals and other arrays and move to single particle methods. A realistic goal is atomic resolution density maps in 24 hours from partially purified preparations. Partially purified preparations of isolated assemblies in frozen suspension would be examined by automated particle identification (purification in silico). Crystallization is bypassed, and even extensive biochemical purification would be unnecessary. In this "blot and automate" approach, 105 particle images in 103 image fields would be recorded over about 8 hours and the data transferred to a teraflop computer for particle selection and 5-D cross-correlation.
Goals for Cells
(1) Conventionally embedded and sectioned cells in 3-D at low resolution by cellular tomography (extension of traditional EM to 3-D). A historical limitation of conventional thin-section EM has been that it produces 2-D projection images of 3-D specimens. The superpositioning of 3-D structural detail in the 2-D image obscures spatial relationships and blurs detail. Modern tomographic methods can produce 3-D images of traditional fixed, stained, embedded, thin-sectioned specimens that are much more informative than 2-D projections. For tomography, the thin-section is tilted in the microscope on a tilt stage, images (typically about 100) are recorded at regularly spaced orientations, and they are computationally combined by 3-D reconstruction. The technique is inherently a low-resolution one. Each specimen is unique, so all the data must be recorded from it. The electron exposure required destroys all of the native chemical structure of the specimen, so it must be chemically treated to convert it into a radiation resistant form before it is analyzed. Despite these limitations, extending the capability of EM to 3-D has the potential for enormous impact in molecular cell biology in the immediate future. Tomographic approaches to conventionally embedded cells will have to serve as a central structural tool in integrative cell biology until high-resolution methods applicable to natively-preserved cells are developed. To realize its potential, cellular tomography must be automated as a high-throughput, low-overhead method.
Supporting specimen technologies needed are: (1) better methods of quick-freezing, fixing, embedding, and staining for more uniform and faithful preservation of specimen detail, (2) methods of rapid freezing to capture transient states, and (3) methods for specific labeling and localization of macromolecules. Supporting instrument technologies that should be exploited are: (1) electron energy filter (for elemental analysis and removal of secondary scattered electrons), (2) high accelerating voltages (for thick specimens), and (3) aberration correctors (for thick specimens).
(2) Native cells. The organizers asked the participants to evaluate prospects for determining macromolecular arrangements in cells.
Rationale: The potential of EM for visualizing macromolecules in native cells has not been realized, but converging advances in structural biology may bring this goal into reach. (a) At the current rate of new structure determination by X-ray and NMR methods, atomic models of many major cellular components will be available in the next decade. New initiatives in protein structure will further increase the pace of discovery. (b) Advances in EM are improving its high-resolution imaging capabilities. (c) More powerful methods of computer processing will be available for extracting high-resolution information from noisy images and for detecting high-resolution signatures of individual macromolecules or assemblies of known structure in images of complex specimens.
The question posed is whether knowledge-based methods can be developed that combine these technologies to define macromolecular 3-D arrangements and interactions in vivo. In outline, possible strategies are: (1) cut sections of natively preserved (e.g., frozen-hydrated) cells thin enough for high-resolution imaging (<100 nm); (2) record limited low-dose tilt data (e.g., stereo image pairs; because of the short, radiation-limited exposure, only incomplete high-resolution detail can be recorded); (3) process the images to remove identifiable noise; (4) detect molecular projection signatures in the images using pattern recognition and a look-up table of known atomic structures of macromolecules and macromolecular assemblies; and (5) triangulate signatures in 3-D (e.g., from tilt images).
Feasibility: This goal of determining macromolecular arrangements in native cells is at least partially achievable. For example, it should be straightforward to detect large assemblies like ribosomes, microtubules, dynein, and dynactin in 50 nm thick frozen sections. The theoretical lower size limit detectable is not known. For individual macromolecules below the lower size limit, tagging will be necessary.
Technologies needed: Preparing sections of natively-preserved cells thin enough for high-resolution imaging (<100 nm) will be a major hurdle. Cells must be mechanically stabilized for sectioning without disrupting native macromolecular arrangements. One approach is to quick-freeze cells and cut thin cryosections in which native cellular structure is preserved. An alternative approach of unknown feasibility (for high resolution) is chemical embedding. Existing fixing and embedding methods are not suitable; new methods would certainly be required.
Entirely new algorithms for noise reduction, data management, and molecular pattern recognition would be required. The knowledge base for look-up will be a comprehensive database of atomic structures of macromolecular assemblies and of individual macromolecules, developed by complementary EM, X-ray and NMR approaches. At current rates of progress, it seems likely that a relatively complete database of atomic structures will be available before the methodology for this goal is perfected.
Session 4: Technology and Technical Problems
Moderator: Bob Glaeser
The purpose of this session was to identify critical technologies needed to achieve the goals and classify the technical problems that must be solved.
Classifications of Technology
Specimen Preparation
Tomography
Improved fixation, staining, embedding
Specific labeling
High resolution
Assemblies: reproducible freezing and cryo-specimen preparation
Natively-preserved sections of cells thinner than 100 nm
(a) Cryosections of frozen-hydrated cells
(b) Sections of embedded natively preserved cells
Specimen-Beam Interaction
Electron Optical Instrumentation
High resolution
Cold stages
Electron source
Cellular tomography
Data Collection
Detector technology
Automation
Computing
Data management
Automation
Image processing and corrections
Specimen variability and classification
Interpretation and model building
Pattern recognition
Knowledge-based methods
Classifications of Difficulty
Alpha
Coming anyway, will be developed in the course of ongoing research, or will be provided by industry.
Beta
Known solution, but implementation requires targeted development efforts by investigators, financial support is necessary.
Omega
Solvable, but the solution is unknown, requires both research and development by investigators, financial support is necessary.
Key to Applications:
(1) Frozen-hydrated arrays and particles at high resolution
(2) Atomic resolution structures of macromolecular assemblies in solution
(3) Conventionally embedded and sectioned cells in 3-D by cellular tomography at low resolution
(4) Macromolecular arrangements in natively-preserved cells in 3-D by EM and knowledge-based approaches
| Technology |
Application |
R&D Category |
|
1 |
2 |
3 |
4 |
|
| Specimen Preparation |
|
|
|
|
|
| Improved methods fixing, embedding, staining |
|
|
X |
|
Acceptable alphaSignificantly improved omega |
| Specific labeling of macro-molecules in solution and in cells |
X |
X |
X |
X |
Acceptable alphaOptimal omega |
| Reproducible, automated cryo-specimen preparation |
X |
X |
|
|
Beta |
| Cryosections thinner than 100 nm |
|
|
|
X |
Assessments varyBeta-omega |
| Embedding and sectioning thinner than 100 nm of natively-preserved cells |
|
|
|
X |
Omega |
| Technology |
Application |
R&D Category |
|
1 |
2 |
3 |
4 |
|
| Beam Specimen Interactions |
|
|
|
|
|
| Radiation damage |
X |
X |
|
X |
alpha |
| Charging |
X |
X |
X |
X |
assessments varyalpha-omega |
| Beam induced movement |
X |
X |
|
X |
assessments varyalpha-omega |
| Technology |
Application |
R&D Category |
|
1 |
2 |
3 |
4 |
|
| Electron OpticalInstrumentation |
|
|
|
|
|
| High resolution |
|
|
|
|
|
| Source coherence |
X |
X |
|
X |
alpha |
| Electron optical performance |
X |
X |
|
X |
OK already |
| Cold stage |
X |
X |
|
X |
acceptable alpha
improved beta |
| Improved CTF correction |
X |
X |
|
X |
beta-omega
(aberration
corrector
omega) |
| Tomography of Convention-ally Prepared Sections at Low Resolution |
|
|
|
|
|
| Tilt stage |
|
|
X |
|
acceptable nowdouble tilt beta |
| Reduced inelastic scattering background |
|
|
X |
|
energy filter beta |
| Technology |
Application |
R&D Category |
|
1 |
2 |
3 |
4 |
|
| Data Collection |
|
|
|
|
|
| Detector Technology |
|
|
|
|
|
| Size |
X |
X |
X |
X |
omega |
| Readout speed |
X |
X |
X |
X |
omega |
| Noise |
X |
X |
X |
X |
omega |
| Electron deceleration and detection |
X |
X |
X |
X |
omega |
| Automation |
|
|
|
|
|
| Operation of controls |
X |
X |
X |
X |
beta |
| Real-time image processing for automatic particle selection |
X |
X |
|
X |
omega |
| Beadless alignment |
|
|
X |
|
beta |
| Remote operation |
X |
X |
X |
X |
beta |
| Intelligent operation |
X |
X |
X |
X |
omega |
| Technology |
Application |
R&D Category |
|
1 |
2 |
3 |
4 |
|
| Computation Algorithm Development |
|
|
|
|
|
| Improved CTF correction |
X |
X |
|
X |
beta-omega |
| Data management |
X |
X |
X |
X |
omega |
| Classification |
X |
X |
X |
X |
omega |
| Pattern recognition |
X |
X |
X |
X |
omega |
| High-resolution model building |
X |
X |
|
X |
beta |
| Tomographic results display |
|
|
X |
|
beta |
| Tomographic results interpretation |
|
|
X |
|
beta |
| User interfaces |
X |
X |
X |
X |
beta |
| Knowledge-based methods |
|
|
|
X |
omega |
Session 5: Solutions
Moderator: Bridget Carragher
Specimen Preparation: Dick McIntosh
Fixation, Embedding, Staining for Cellular Tomography at Low Resolution
Application: Cellular tomography at low resolution only, not relevant for high resolution.
Solutions: Better fixatives, water-soluble embedding agents, better stains, low temperature freeze substitution.
Current fixing, embedding and staining methods were developed in the 1950s and are showing their age. Uniformity is poor, and the chemicals disrupt native structure. Better chemicals and methods are needed for conventional work. Fast freezing is accepted as the best way to begin because all molecules in the specimen are immobilized rapidly, regardless of their chemistry. The retention of high levels of enzyme activity, and even of cell viability, through fast freezing encourages confidence in this method. With current methods, one transfers a frozen cell to a liquid solvent at low temperature, so the cell's water will dissolve and allow some "fixation" to stabilize the structure for further study (freeze-substitution fixation). New fixation protocols should improve specimen preservation for imaging in plastics at 3-6 nm resolution, also permitting tagging with specific labels. Eventually, if the capability to analyze cryosections of frozen-hydrated cells can be developed (Goal 4), chemical fixation, embedding, and staining may be avoided altogether.
Specific Labeling for Identification of Macromolecules in 3-D Maps
Applications: Cellular tomography at low resolution, natively preserved cells at high resolution, large isolated macromolecular assemblies.
Solutions: Antibody tags, chimeras, direct labeling.
Approaches are: (1) antibodies, using epitope labels and (2) chimeras (fusions with tags like the green fluorescent protein). Novel tags may bind enough copies of a unique atom to become visible directly, and the conversion of fluorescent tags to EM-visible stains has been achieved in some cases. These methods must be modified to work at low temperature on freeze-substituted specimens. This is a more difficult task, but one that should be accomplished within 5 years. Work on these methods will probably fall into two categories: (1) finding tags with high signal to noise that allow the identification of rare molecules distributed over extensive areas, e.g., a cisterna in the Golgi apparatus; and (2) finding tags that can be used for higher resolution work in dense structures, like a kinetochore. Progress here should be obvious within 5 years, providing additional motivation for the harder job of imaging natively preserved cells with sufficient resolution to identify macromolecules directly.
High Resolution
Reproducible Freezing and Cryo-Specimen Preparation
Applications: Isolated frozen-hydrated macromolecular assemblies at high-resolution
Solution: Automation
Cryo-specimen preparation is much more of a craft than a science. Currently, it requires a high degree of skill. Exquisite timing and dexterity are required to precisely control parameters like ice film thickness. Instruments are needed that precisely control all variables, according to settings dialed in by comparatively unskilled operators. To develop them, collaborative mechanical engineering expertise should be enlisted, probably from industry.
Cryo-Sections Thinner Than 100 nm
Application: Intact frozen-hydrated cells at high resolution
Solution: Cryo-ultramicrotomy of frozen cells
For the determination of macromolecular arrangements in native cells, thin-sections of cells that are 100 nm thick or less are required. Cells must be mechanically stabilized for sectioning without disrupting native macromolecular arrangements. One approach is quick-freezing and cryosectioning of unfixed, unstained cells. Assessments of difficulty and prospects varied; after the meeting we consulted Dr. Ken Taylor (Florida State University), who wrote:
"I believe that it is possible to cut 100 nm thick sections of unfixed material, and if you keep it cold, you can look at it frozen hydrated. Fifty nm thick sections would not be a technical tour-de-force, but would be demanding. Not just anyone could cut them. I think that the prevailing wisdom on frozen thin sectioning is that it is mostly freeze fracture, but you can still cut something thin."
Embedding Methods
Application: Intact natively preserved cells at high resolution
Possible solution: An alternative approach of unknown feasibility (for high resolution) is chemical embedding. Perhaps methods and reagents can be developed to mechanically stabilize cells for thin-sectioning, yet preserve macromolecular arrangements.
Research and Development Strategies
Extensive collaborations with outside experts (chemists for improved fixing, embedding, staining, and labeling and mechanical engineers and materials scientists for instrument design and automation) are needed for faster progress in automating and improving specimen technologies.
Specimen-Beam Interactions: Ken Downing
Radiation Damage
Application: High resolution
Solution: Liquid helium stages (alpha, possibly beta)
Scattering of the electron beam by the specimen provides all of the information used to produce an image or diffraction pattern. Elastic scattering provides the information used in molecular studies, while inelastic scattering causes several problems that interfere with the information. Radiation damage is the most serious of all problems in biological studies, limiting the exposure that specimens can tolerate. This in turn limits the signal-to-noise ratio that can be achieved and thereby limits the resolution. Small radiolytic fragments are produced that can diffuse around within the specimen and evaporate from the surface, leading to specimen motion and mass loss. The inelastically scattered electrons produce a haze that decreases image contrast. Depending on the specimen thickness, electrons may be scattered more than once, elastically, inelastically, or both. Further, some of the inelastic scattering causes secondary electrons to be ejected from the specimen, leading to specimen charging, which then causes beam and image deflections that can seriously reduce resolution. We can limit the events that follow initial damage in the specimen. Low temperatures limit fragment diffusion. Specimens cooled by liquid nitrogen last as much as 10 times longer in the beam than when at room temperature. There may be around another factor of two in the lifetime of crystalline specimens at helium temperatures, and beam-induced specimen motion may be significantly reduced by cooling below nitrogen temperature, an effect that requires more extensive investigation. Continuing advances in cold stage design to bring the temperature of the specimen closer to that of the coolant (from 20°K to closer to 4°K) should make possible further improvement.
Charging
Applications: All
Charging is particularly serious with plastic sections and ice, since they are such good insulators, but can even be seen on many samples supported on continuous carbon films. The problem is more acute when the specimen is tilted. Remedies of varying complexity were discussed.
Alpha solutions: Among the most effective ways to reduce effects of charging so far is having part of the electron beam strike a surface that can induce sufficient emission of secondary electrons to neutralize the charge on the specimen. The holey carbon around an ice-embedded specimen or the objective aperture are both sources of secondary electrons. Increasing the secondary electron emission coefficient of either could provide a larger electron source for neutralizing charge. It is likely to be essential to avoid any ice condensation on the specimen, as this would produce an insulating surface subject to charging. Along these lines, Dr. Unwin discussed three empirical methods for reducing charging effects: elimination of aperture contamination, beam-hole centering, and pre-irradiation of support films. Specifically, these were: (1) ultra-cleaning of objective apertures, (2) centering the beam over holes in the support film and adjusting its width to match or exceed the hole diameter, and (3) pre-irradiation of the carbon film in high vacuum, making it 20 times more conductive. Dr. Unwin said that in the case of acetylcholine receptor, these precautions eliminated all symptoms of charging. One school of thought held that careful adjustments to normal operating procedures like these would eliminate the problem for untilted specimens.
Beta solutions: Conductive support films and specimen coating. (1) Coating samples with amorphous, high-conductance materials would increase conductivity, but obvious candidate materials are grainy and would obscure specimen detail. (2) The conductivity of carbon support films decreases with temperature. Alternatives are alloys or conductive polymers.
Omega solutions: low energy ion sources near specimen and/or objective aperture.
Research and Development Strategies
New approaches should be tested with a sufficient number of specimens and images to determine both the improvement in image quality and the improvement in the success rate of obtaining good images. In general, one needs to quantify the falloff with increasing resolution of amplitudes in the Fourier transform of the image. Automation of specimen identification and data collection and processing would speed up the evaluation process dramatically.
Electron Optical Instrumentation: Wah Chiu
Optimal instrument configurations for high resolution and tomographic work differently and are treated separately.
High Resolution
The resolving power of commercially available transmission electron microscopes is better than 2.5 Å. Resolutions below 2Å have been routine in materials science for years. For specimens under 100 nm thickness at the highest resolutions, electron optical performance is already fully adequate and signal loss due to aberrations is not significant (note, however, that CTF correction is a major issue).
Biological macromolecules are relatively large and very radiation sensitive. Special features in an electron microscope are necessary to realize its imaging potential for biological macromolecules at atomic resolution. These features include:
- 200-400 keV electron beam energy, which has less chromatic aberration and a larger depth of field.
- A field emission gun, which is a bright and coherent electron source to record images with high resolution and high contrast. Imperfect coherence of the electron source results in loss of signal when the elastically scattered electrons are combined to form the image. It is anticipated that continuing improvements in gun design will further improve coherence.
- A specimen holder operating at or below liquid helium temperature to keep specimen frozen and hydrated and to reduce radiation damage.
- A 2k x 2k CCD camera, which allows online digital data collection and evaluation.
The current cost of all these features is about $2-2.5 million. Typical site preparation costs are $700 thousand. Total costs are about $3 million per installation.
Current approaches to correcting for defocus and other components of the CTF are not good enough to support high-throughput data collection at high resolution. Furthermore, current methods are adequate at high resolution (3.5 Å) only for very thin specimens (under 50 nm). Improvements are necessary if thicker specimens (50-100 nm) are to be analyzed at high resolution. Extensive work is needed to improve experimental procedures and algorithms for image processing. Also, promising electron optical innovations have the potential to simplify CTF correction, (e.g., aberration correctors, which can modify the CTF) but will require extensive development and evaluation.
Cellular Tomography at Low Resolution
The ability to tilt the specimens at high angles for 3-D data collection is required. New features are moving to market that should further enhance data quality and collection efficiency. They include: (1) post-specimen energy filter to enhance the image contrast, (2) large format electronic cameras with an improved MTF and S/N, and (3) higher accelerating voltages (1,000 keV) to image thicker specimens.
Accelerating Voltage. For tomographic analysis of thick specimens (over 200 nm thickness) at lower resolution, higher voltages (1 MEV=1000 kV) are better. Note, for high-resolution work, including cryo-sections of frozen-hydrated cells, which are thin specimens, 300-400 kV appears appropriate, but more work is needed to define optimal conditions.
Energy Filter. The energy filter removes inelastically and secondarily scattered electrons, reducing background noise in the image.
Signal Losses in Electron Microscopy Systems
Net signal loss in the entire EM system may be approximated by the envelope decay function. Major contributing factors are imperfect spatial and temporal source coherence, detector deficiencies (imperfect detector modulation transfer function, MTF), inelastic scattering, radiation damage, beam-induced specimen movement, charging effects, and stage instability. For high-resolution work, objective lens aberrations are not a significant source of signal loss, even at 2.5Å resolution and beyond. However, as noted above, correction of defocus and other components of the CTF is a major technical issue at high resolution.
Each of the contributing factors to signal loss has its own envelope function. The overall decay function is the product of the different envelope functions and may be approximated as a Gaussian function:
Where D is the net envelope decay factor, B is the amplitude decay and S is the spatial frequency (reciprocal of the resolution in Å). D describes signal fall-off as a function of resolution. B is the useful single number that describes the performance of an entire microscope system, including image recording and digitization.
If the EM system performs perfectly, its amplitude decay (B) is equal to zero, and its envelope decay function (D) is equal to 1.0 (100 percent signal recovered) at all resolutions. The smaller the amplitude decay (B), the better the microscope system.
Current images from the best 200-300 kV microscopes have B=30-60Å2. For B=30Å2, D=0.3 at 4 Å resolution and D = 0.15 at 3 Å resolution. There is scope for three to six fold improvement. The largest factors contributing to envelope decay are still source coherence and detector MTF. Improved FEGs and detectors should lower B further.
Research and Development Strategies
Close cooperation with industry speeds the introduction of improved technologies. More work is needed to evaluate innovations in electron optical instrumentation that may improve performance or simplify problems with data collection. Aspects requiring more study are accelerating voltage, the use of energy filters, and applications of aberration correctors, which may simplify CTF corrections to image data. There is a need for better ways to benchmark the performance of EM components. Artificial radiation-insensitive test specimens would be a major asset.
The demands for near-atomic to atomic resolution EM analysis of single particles place extraordinary burdens on instrumentation, data collection, and computation. Current calculations suggest that on the order of 105 single particle images will need to be recorded and combined to generate a high-resolution 3-D reconstruction of the object. To obtain a resolution of 3Å requires pixels of about 1Å. For a 200 Å diameter structure, this translates into recording about 5,000 2Kx2K pixel images. For a 500 Å structure, the requirement is about 25,000 2Kx2K images or about 6000 4Kx4K pixel images. While EM tomography does not require so many images, examining cellular structures requires 6-7Å pixels, which for objects in the size range of one to five microns demands detectors having from 2Kx2K to 10Kx10K pixels.
While today's high-resolution work employs film, the practicality of dealing with so many images dictates that digital image data be obtained online. Also, immediate access to digital images is an absolute prerequisite for the automation described below. Although research efforts are underway to develop alternative detectors, for the near future, (next 5 years) charge coupled device CCD detectors are the best devices available.
Problem: Detectors
Problems with current CCD detectors at about 300 keV include: limited size (1K2 or 2K2), poor stopping quantum efficiency for phosphor at about 300 keV, poor match between phosphor modulation transfer function (MTF) and CCD pixels (true pixel size is about 2 times physical pixel size leading to smaller effective image size), slow readout speed, high readout noise, and a serious additional noise due to backscatter at higher acceleration voltages (a large fraction of the electrons pass through the phosphor and enter the fiber optic where they can be scattered back to the phosphor, adding long tails to the detector point spread function or PSF).
Developing improved phosphors optimized for 300keV electrons will be quite important. Large area CCD detectors with pixel sizes that are properly matched to the point spread function of the phosphor are required. In addition, readout speeds need to be significantly improved and readout noise needs to be decreased. Solving the backscatter problem is critical for low-noise, high-resolution imaging. Two choices are to use either a lens coupled system to avoid the fiber optic or fancy electron optics similar to that used in imaging energy filters to slow down the electrons. This would allow the electrons to be completely captured by the phosphor, or in the extreme, permit direct imaging of incident electrons right on the CCD.
Problem: Automation
Fully automating data collection is critical. Automatic microscope setup should be followed by automatic location of areas of suitable ice thickness within holes at low magnification. At intermediate magnification, image areas containing suitable numbers of particles can be recognized and then data collected at high magnification. Auto positioning must be an integral aspect of data collection. These requirements impose enormous real-time computational demands.
Research and Development Strategies
Extensive collaboration with scientists and engineers in other fields will be essential. In astronomy, the state of the art in detector technology is far more advanced than in microscopy.
Classes of Specimen
In 3-D imaging with EM, computational challenges are different for the different classes of specimens ranging from (1) 2-D crystals, (2) viruses with icosahedral symmetry, (3) fibers and assemblies with helical symmetry, (4) asymmetric particles, and (5) "bulk" sections or whole cells.
Volume of Data
There is a roughly cubic dependence between the volume of data to be collected and processed and the resolution. This dependency allows us to estimate the trend in the need for computational resources, as resolution is being pushed for all classes of specimens, and compare it with the "natural" trend in the commercial development of computer hardware.
Symmetrical and Periodic Structures
Specimen variability greatly complicates all computational approaches designed to extract an average, high-resolution structure (specimens of classes 1-4). In the case of symmetrical structures (1-3), the presence of any disorder counteracts the assumption of the underlying symmetry. For these classes of specimens, a major part of all computational resources (algorithm design, programming, and CPU use) goes into efforts to restore the order. There is the consensus that major hurdles have been overcome through the development of unbending techniques (including the use of a quasi-single particle approach) and various methods of refinement.
Asymmetric Particles and Classification Methods
For single particle specimens without any symmetry (class 4), the problem of disorder appears in an entirely different form: the complete absence of neighbor constraints allows the macromolecular assembly to assume a wide range of conformational states. This is both a curse and an opportunity. It is a curse because it makes the data collection cumbersome--a very large data set has to be collected to fulfill the statistical requirements for any selected conformational state--and because the methods for screening out a homogeneous subset from a heterogeneous population of molecules are far from settled. It is an opportunity, on the other hand, because it allows the sampling of functionally potentially meaningful conformational space. Here, the investment must be in statistical methods that allow rigorous "multireference" methods to be developed and validated. It may be necessary to use multiple very low-dose tomographic reconstructions of the same macromolecule for validation.
Unique Structures
3-D imaging of bulk material by electron tomography (class 5), e.g., 3-D imaging of mitochondria, cannot make use of averaging, so specimen variability does not pose a computational problem--it only poses a heuristical problem, since the question is to what extent the result typifies the particular class of structures studied.
Contrast Transfer Function Correction
A common element in all image processing arises from the fact that all data originate from the electron microscope, which has a unique image formation characteristic, expressed (in approximation) by the contrast transfer function, and a fall-off of information toward higher resolution that is presently not well understood. Investment in both experimental and theoretical research of these issues, along with development of algorithms of CTF correction that are closer to the physical reality, is a must if we wish to approach atomic resolution more routinely.
Some of this information will come from a meticulous comparison of 3-D maps that have been obtained by both cryo-EM and X-ray crystallography, so it is desirable, for the good of the whole field, to sponsor a selected number of such comparative studies, even if no new structural information will ensue.
Interpretation and Model Building
Interpretation involves a variety of visualization tools that allow the EM density map to be displayed jointly with crystallographic data. There is a great need in this area for the development of tools that simplify, guide, and possibly automate the job of fitting and docking NMR and X-ray structures into cryo-EM density maps. Commercial developments, such as those used for computer-guided surgery, and in general, a much stronger involvement of computer graphics experts, would be extremely helpful in this area.
Tomographic 3-D Reconstruction
Problems unique to class 5 specimens, which are imaged using tomographic tilt series, are the need to develop efficient and robust algorithms for projection alignment without markers (beads) and the need for improved reconstruction algorithms. The push toward larger areas, thicker sections, and improved resolution for such specimens will result in 3-D arrays of unprecedented size, with processing, visualization, and "mining" that poses entirely new challenges that might require use of new technologies of accessing and storage. The display, interpretation, and communication of 3-D tomographic results pose entirely new challenges.
Automation
The computer actually comes into play in EM structure research in several ways. So far, I have dealt with the number-crunching role only. The other roles, which are of increasing importance, are in the automation of data collection and in its interpretation. It should be stressed that the ancillary role of the computer in the data collection will be extremely important for achieving our goal. Very powerful hardware will be needed.
Molecular Signatures
For knowledge-based approaches to image processing that would allow molecular projection signatures to be identified, we have to envision some type of screening that makes use of 3-D data residing in locally, as well as publicly accessible, databases. Algorithm development for pattern recognition is an area that will benefit from interaction with experts in a variety of areas of image processing that are normally untapped.
User Interfaces and Documentation
In a structural biology group, the computer is the platform that brings together people from many different backgrounds. Not only the state of algorithm development, but also the ease with which software can be used by non-experts, is crucial for efficiency in research. One aspect is the soundness, completeness, and intelligibility of the documentation. Another is the friendliness of the user interface. The rapid development of hardware and software platforms has hopelessly eclipsed the specialized software packages that were developed by academic groups in the 1980s. It is necessary to invest resources (programmer positions, consultations, interdisciplinary collaborations) in this area to close the widening gap and to keep the gap from opening again in the future.
Research and Development Strategies
Personnel
There is a general point that needs to be made. There has been a reluctance of the existing funding mechanism (within R01s at least) to allow for computer specialists, even though they are often proposed in the context of cutting-edge algorithm and methods development within a relevant testbed. I would suggest that proposals in the areas of EM-related structural biology should be carefully checked for opportunities of pilot developments that have the potential to benefit the entire field.
Training
To develop new scientists with the requisite computational skills, strong interdisciplinary training will be necessary, particularly in conjunction with fields of mathematics and computer science.
Expert Collaborators
Collaboration with computer scientists and mathematicians on problems of data management, image processing, pattern recognition and classification, noise treatment, graphics, expert system design, and automation will be required.
(1) R01 support levels are good. High-resolution EM research grant applications on significant topics are enthusiastically received by NIH peer reviewers and enjoy high success rates.
(2) There is a strong core (about 20) of active and productive senior PIs. After a long hiatus, a number of outstanding new investigators (up to 10) have recently been hired into independent positions at leading institutions. A number of leading institutions are recruiting faculty in this area.
(3) There has been a marked increase in interest in the field, evidenced by high profile recruiting by leading institutions. Private foundations that have recently directed resources into the field include HHMI, the Agouron Foundation, and the Keck Foundation.
(4) There have been some spectacular recent accomplishments. Two recent high-profile examples are the atomic structure of tubulin and the 9 Å structure of the ribosome by combined crystallographic and electron microscopic approaches.
(5) New areas and technologies are being explored and opened. Examples include single-particle methods, networking for remote operation and data acquisition, automation, detector technology, new methods for cryospecimen preparation, and new electron-optical technology.
(6) Resolution has been advancing at a steady pace, with several examples of atomic resolution on 2-D crystals, 7-9 Å resolution on specimens of lower redundancy like viruses, and 15 Å on a asymmetric particle, the ribosome.
(1) Relative to the fields of macromolecular X-ray crystallography and NMR, the field of high-resolution EM in the United States is small, with fewer than 30 active groups. Despite recent high profile recruiting, the field is understaffed relative to the scientific challenges.
(2) Much of the research support comes from R01s built around biological specific aims. Consequentially, most of the research effort is directed toward short-term goals. Not all, however. For example, the SPIDER system and single particle 3-D reconstruction were developed in the Frank lab over a period of two decades with short-term NIH support.
(3) There is considerable duplication of effort. Only a few periodic specimens are both important and offer prospects of high-enough resolution to support grant applications and publications. As a consequence, groups in the United States and abroad often compete on the same problem (for example, four groups working on kinesin-decorated microtubules, two or three groups on the same virus, four groups on decorated actin filaments, two groups on ryanodine receptors, etc.).
(4) Many obvious technical problems have not been addressed by directed, systematic efforts. Most efforts in technology development are on a part-time, shoestring basis, supported by grants nominally awarded to analyze specific structures. PIs noted that study sections routinely cut personnel designated for development of methods specimen preparation or image processing.
(5) The technology is underdeveloped and the need for improvement is widely recognized; yet there are few applications directed toward improving methodology. This is in striking contrast to the situation in NMR, where NIH supports a very robust program of technology R&D. One factor seems to be the perception that the review system is hostile to R&D. Another may be lack of the expertise in engineering and the hard sciences needed to tackle some of the most difficult problems.
(6) Most of the investigators in the field have similar skill sets. The PIs tend to be jacks of all trades because of the range of skills needed to carry a project through from the biochemistry to the image processing. However, this breadth and versatility comes at the expense of depth in the critical areas of mathematics, engineering, and physics. Aggregate in-house expertise in these fields is much more limited than in the fields of crystallography and NMR. It's not clear that the needed skills are available at the entry level, or that existing training mechanisms in biophysics would produce scientists that have the skills that are most needed.
(7) Although there is a clear need and numerous opportunities to contribute, there is no obvious gateway through which established physicists, mathematicians, and engineers can enter EM from other fields as funded investigators.
(8) The experimental work is often repetitive, requiring painstaking effort sustained through many unsuccessful repetitions before a positive outcome is achieved. A high degree of manual dexterity is required for many basic operations, and it takes years to develop the skills needed. The tedium discourages entry of young scientists into the field. One contributing factor is dependence on large, perfect, periodic specimens, which are delicate and rare. Other related problems are lack of automation and the still generally primitive state of the art (for example, the lack of adequate electronic detectors makes necessary the recording, processing, and analysis of hundreds or thousands of photographic films).
(9) In addition to the difficulties noted with entry-level personnel, there are also problems with retention of skilled and experienced personnel over the long-term. It is difficult to raise the higher salaries needed for senior, non-tenure track research associates on research grants, and this support is subject to the interruptions and vagaries of the peer review system.
(10) Many PIs are working with obsolete microscopes, which limits their progress and the quality and impact of their work. To acquire a modern 300 KV FEG liqHe microscope with associated accessories, space, and renovations, most PIs would have to orchestrate support from public, private, and institutional sources. The only successful acquisition to date through agency sources required simultaneous applications to NSF, the Keck Foundation, local foundations, and contributions from the department and various deans. Many PIs are superb scientists, but lack the knowledge, patience, political acumen, and time needed to orchestrate such a complex funding effort.
(11) Conclusion: The format of the review and funding systems do influence what research is done and how it is conducted. Because of the pressure to produce incremental progress on biology over the term of a 4-year funding cycle, it appears that important long-term technical development work has been neglected in favor of shorter-term, sometimes redundant, efforts to produce information of immediate biological significance.
The standard in-house microscope for routine work, specimen work-up, refinement of procedures, etc., is now a 200 kV with FEG. To be competitive, new investigators need daily access to an instrument of this caliber. A number of established investigators are using obsolete microscopes and need to upgrade.
For state of the art work, a 300 kV, FEG, liqHe instrument is essential. These should be shared by groups of investigators and operated continuously.
Anticipated computing demands will be three to four orders of magnitude greater than present, so significant investment in computing hardware will be necessary.
High-caliber, career-track support personnel are needed to maintain 300 KV microscopes in peak operating condition with round-the-clock availability to users.
As the sophistication of the instrumentation increases, more support personnel will be needed in individual laboratories.
With the transition to automation and high-throughput operation, more programmers will be needed for software maintenance and development.
Support for R&D projects targeted to hardware and software development is essential. Appropriate peer review arrangements should be made for R&D applications.
Funding opportunities should be created for investigators from the fields of physics, mathematics, materials science, engineering, and computer science for collaborative R&D projects (for example, "glue grants").
Outreach to scientists in other fields is required to recruit needed expertise. One recruiting approach is to organize workshops or courses to publicize research opportunities and invite established scientists and engineers from other fields.
Interdisciplinary graduate and postdoctoral training is needed.
Development of improved EM capabilities is critical for optimal progress in emerging integrative research in molecular cell biology, which depends on a structural approach in conjunction with genomics to defining the functions of molecular machines and networks.
Ten to 20 years of concentrated effort on technology development may be needed to realize the practical potential of EM in biology. Long before then, the underdevelopment of the technology will be an expensive bottleneck. Until the capability for direct visualization of macromolecular arrangements in very large assemblies and intact cells is developed, indirect approaches that are much slower and more expensive will have to be customized for each class of problems. This will delay the pace of progress in many important areas of integrative cell biology. While these capabilities are under development, transitional technologies, in particular cellular tomography, should be developed for high-throughput so they can be applied as routine structural tools.
Equipment and support. A top priority is acquisition of the next generation of electron microscopes and support for their operation, especially high-caliber personnel.
Technology development. Opportunities for support for technology development should be publicized. Although the participants who attended this workshop now have a much better picture of the opportunities for flexible support, the broad community does not. An announcement in the NIH Guide of interest in technology development to improve EM capabilities would stimulate research, help recruit senior collaborators from other fields, have a favorable impact on review, and facilitate negotiation for institutional cofunding and infrastructure support.
David Agard (University of California, San Francisco)
Pamela Bjorkman (California Institute of Technology)
Bridget Carragher (Beckman Institute, University of Illinois atUrbana-Champaign)
Wah Chiu, Chair (Baylor College of Medicine)
David Davies (National Institutes of Health)
David DeRosier (Brandeis University)
Ken Downing (University of California, Berkeley)
Joachim Frank (Wadsworth Center, N.Y. State Department of Health)
Bob Glaeser (University of California, Berkeley)
Steve Harrison, Chair (Harvard University)
Jim Hurley (National Institutes of Health)
Jennifer Lippincott-Schwartz (National Institutes of Health)
Paul Matsudaira (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology)
Dick McIntosh (University of Colorado)
Ron Milligan (The Scripps Research Institute)
Mary Porter (University of Minnesota)
Alasdair Steven (National Institutes of Health)
Susan Taylor (University of California, San Diego)
Nigel Unwin (Medical Research Council, Laboratory of Molecular Biology)
Please direct inquiries to:
James F. Deatherage, Ph.D.
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
NIH Building 45, Room 2AS.13J
Bethesda, MD 20892-6200
Tel: (301) 594-3828
Fax: (301) 480-2004
E-mail: deatherj@nigms.nih.gov
Ms. Teresa Trent, who made all of the administrative arrangements and ran the meeting.
Dr. James Cassatt, who provided support and guidance.
Dr. Jean Chin, who assisted with the running of the meeting.
Drs. Jean Chin, Catherine Lewis, Sue Shafer, and Don Schneider, who took notes.
he backscatter problem is critical for low-noise, high-resolution imaging. Two choices are to use either a lens coupled system to avoid the fiber optic or fancy electron optics similar to that used in imaging energy filters to slow down the electrons. This would allow the electrons to be completely captured by the phosphor, or in the extreme, permit direct imaging of incident electrons right on the CCD.
Problem: Automation
Fully automating data collection is critical. Automatic microscope setup should be followed by automatic location of areas of suitable ice thickness within holes at low magnification. At intermediate magnification, image areas containing suitable numbers of particles can be recognized and then data collected at high magnification. Auto positioning must be an integral aspect of data collection. These requirements impose enormous real-time computational demands.
Research and Development Strategies
Extensive collaboration with scientists and engineers in other fields will be essential. In astronomy, the state of the art in detector technology is far more advanced than in microscopy.
Classes of Specimen
In 3-D imaging with EM, computational challenges are different for the different classes of specimens ranging from (1) 2-D crystals, (2) viruses with icosahedral symmetry, (3) fibers and assemblies with helical symmetry, (4) asymmetric particles, and (5) "bulk" sections or whole cells.
Volume of Data
There is a roughly cubic dependence between the volume of data to be collected and processed and the resolution. This dependency allows us to estimate the trend in the need for computational resources, as resolution is being pushed for all classes of specimens, and compare it with the "natural" trend in the commercial development of computer hardware.
Symmetrical and Periodic Structures
Specimen variability greatly complicates all computational approaches designed to extract an average, high-resolution structure (specimens of classes 1-4). In the case of symmetrical structures (1-3), the presence of any disorder counteracts the assumption of the underlying symmetry. For these classes of specimens, a major part of all computational resources (algorithm design, programming, and CPU use) goes into efforts to restore the order. There is the consensus that major hurdles have been overcome through the development of unbending techniques (including the use of a quasi-single particle approach) and various methods of refinement.
Asymmetric Particles and Classification Methods
For single particle specimens without any symmetry (class 4), the problem of disorder appears in an entirely different form: the complete absence of neighbor constraints allows the macromolecular assembly to assume a wide range of conformational states. This is both a curse and an opportunity. It is a curse because it makes the data collection cumbersome--a very large data set has to be collected to fulfill the statistical requirements for any selected conformational state--and because the methods for screening out a homogeneous subset from a heterogeneous population of molecules are far from settled. It is an opportunity, on the other hand, because it allows the sampling of functionally potentially meaningful conformational space. Here, the investment must be in statistical methods that allow rigorous "multireference" methods to be developed and validated. It may be necessary to use multiple very low-dose tomographic reconstructions of the same macromolecule for validation.
Unique Structures
3-D imaging of bulk material by electron tomography (class 5), e.g., 3-D imaging of mitochondria, cannot make use of averaging, so specimen variability does not pose a computational problem--it only poses a heuristical problem, since the question is to what extent the result typifies the particular class of structures studied.
Contrast Transfer Function Correction
A common element in all image processing arises from the fact that all data originate from the electron microscope, which has a unique image formation characteristic, expressed (in approximation) by the contrast transfer function, and a fall-off of information toward higher resolution that is presently not well understood. Investment in both experimental and theoretical research of these issues, along with development of algorithms of CTF correction that are closer to the physical reality, is a must if we wish to approach atomic resolution more routinely.
Some of this information will come from a meticulous comparison of 3-D maps that have been obtained by both cryo-EM and X-ray crystallography, so it is desirable, for the good of the whole field, to sponsor a selected number of such comparative studies, even if no new structural information will ensue.
Interpretation and Model Building
Interpretation involves a variety of visualization tools that allow the EM density map to be displayed jointly with crystallographic data. There is a great need in this area for the development of tools that simplify, guide, and possibly automate the job of fitting and docking NMR and X-ray structures into cryo-EM density maps. Commercial developments, such as those used for computer-guided surgery, and in general, a much stronger involvement of computer graphics experts, would be extremely helpful in this area.
Tomographic 3-D Reconstruction
Problems unique to class 5 specimens, which are imaged using tomographic tilt series, are the need to develop efficient and robust algorithms for projection alignment without markers (beads) and the need for improved reconstruction algorithms. The push toward larger areas, thicker sections, and improved resolution for such specimens will result in 3-D arrays of unprecedented size, with processing, visualization, and "mining" that poses entirely new challenges that might require use of new technologies of accessing and storage. The display, interpretation, and communication of 3-D tomographic results pose entirely new challenges.
Automation
The computer actually comes into play in EM structure research in several ways. So far, I have dealt with the number-crunching role only. The other roles, which are of increasing importance, are in the automation of data collection and in its interpretation. It should be stressed that the ancillary role of the computer in the data collection will be extremely important for achieving our goal. Very powerful hardware will be needed.
Molecular Signatures
For knowledge-based approaches to image processing that would allow molecular projection signatures to be identified, we have to envision some type of screening that makes use of 3-D data residing in locally, as well as publicly accessible, databases. Algorithm development for pattern recognition is an area that will benefit from interaction with experts in a variety of areas of image processing that are normally untapped.
User Interfaces and Documentation
In a structural biology group, the computer is the platform that brings together people from many different backgrounds. Not only the state of algorithm development, but also the ease with which software can be used by non-experts, is crucial for efficiency in research. One aspect is the soundness, completeness, and intelligibility of the documentation. Another is the friendliness of the user interface. The rapid development of hardware and software platforms has hopelessly eclipsed the specialized software packages that were developed by academic groups in the 1980s. It is necessary to invest resources (programmer positions, consultations, interdisciplinary collaborations) in this area to close the widening gap and to keep the gap from opening again in the future.
Research and Development Strategies
Personnel
There is a general point that needs to be made. There has been a reluctance of the existing funding mechanism (within R01s at least) to allow for computer specialists, even though they are often proposed in the context of cutting-edge algorithm and methods development within a relevant testbed. I would suggest that proposals in the areas of EM-related structural biology should be carefully checked for opportunities of pilot developments that have the potential to benefit the entire field.
Training
To develop new scientists with the requisite computational skills, strong interdisciplinary training will be necessary, particularly in conjunction with fields of mathematics and computer science.
Expert Collaborators
Collaboration with computer scientists and mathematicians on problems of data management, image processing, pattern recognition and classification, noise treatment, graphics, expert system design, and automation will be required.
(1) R01 support levels are good. High-resolution EM research grant applications on significant topics are enthusiastically received by NIH peer reviewers and enjoy high success rates.
(2) There is a strong core (about 20) of active and productive senior PIs. After a long hiatus, a number of outstanding new investigators (up to 10) have recently been hired into independent positions at leading institutions. A number of leading institutions are recruiting faculty in this area.
(3) There has been a marked increase in interest in the field, evidenced by high profile recruiting by leading institutions. Private foundations that have recently directed resources into the field include HHMI, the Agouron Foundation, and the Keck Foundation.
(4) There have been some spectacular recent accomplishments. Two recent high-profile examples are the atomic structure of tubulin and the 9 Å structure of the ribosome by combined crystallographic and electron microscopic approaches.
(5) New areas and technologies are being explored and opened. Examples include single-particle methods, networking for remote operation and data acquisition, automation, detector technology, new methods for cryospecimen preparation, and new electron-optical technology.
(6) Resolution has been advancing at a steady pace, with several examples of atomic resolution on 2-D crystals, 7-9 Å resolution on specimens of lower redundancy like viruses, and 15 Å on a asymmetric particle, the ribosome.
(1) Relative to the fields of macromolecular X-ray crystallography and NMR, the field of high-resolution EM in the United States is small, with fewer than 30 active groups. Despite recent high profile recruiting, the field is understaffed relative to the scientific challenges.
(2) Much of the research support comes from R01s built around biological specific aims. Consequentially, most of the research effort is directed toward short-term goals. Not all, however. For example, the SPIDER system and single particle 3-D reconstruction were developed in the Frank lab over a period of two decades with short-term NIH support.
(3) There is considerable duplication of effort. Only a few periodic specimens are both important and offer prospects of high-enough resolution to support grant applications and publications. As a consequence, groups in the United States and abroad often compete on the same problem (for example, four groups working on kinesin-decorated microtubules, two or three groups on the same virus, four groups on decorated actin filaments, two groups on ryanodine receptors, etc.).
(4) Many obvious technical problems have not been addressed by directed, systematic efforts. Most efforts in technology development are on a part-time, shoestring basis, supported by grants nominally awarded to analyze specific structures. PIs noted that study sections routinely cut personnel designated for development of methods specimen preparation or image processing.
(5) The technology is underdeveloped and the need for improvement is widely recognized; yet there are few applications directed toward improving methodology. This is in striking contrast to the situation in NMR, where NIH supports a very robust program of technology R&D. One factor seems to be the perception that the review system is hostile to R&D. Another may be lack of the expertise in engineering and the hard sciences needed to tackle some of the most difficult problems.
(6) Most of the investigators in the field have similar skill sets. The PIs tend to be jacks of all trades because of the range of skills needed to carry a project through from the biochemistry to the image processing. However, this breadth and versatility comes at the expense of depth in the critical areas of mathematics, engineering, and physics. Aggregate in-house expertise in these fields is much more limited than in the fields of crystallography and NMR. It's not clear that the needed skills are available at the entry level, or that existing training mechanisms in biophysics would produce scientists that have the skills that are most needed.
(7) Although there is a clear need and numerous opportunities to contribute, there is no obvious gateway through which established physicists, mathematicians, and engineers can enter EM from other fields as funded investigators.
(8) The experimental work is often repetitive, requiring painstaking effort sustained through many unsuccessful repetitions before a positive outcome is achieved. A high degree of manual dexterity is required for many basic operations, and it takes years to develop the skills needed. The tedium discourages entry of young scientists into the field. One contributing factor is dependence on large, perfect, periodic specimens, which are delicate and rare. Other related problems are lack of automation and the still generally primitive state of the art (for example, the lack of adequate electronic detectors makes necessary the recording, processing, and analysis of hundreds or thousands of photographic films).
(9) In addition to the difficulties noted with entry-level personnel, there are also problems with retention of skilled and experienced personnel over the long-term. It is difficult to raise the higher salaries needed for senior, non-tenure track research associates on research grants, and this support is subject to the interruptions and vagaries of the peer review system.
(10) Many PIs are working with obsolete microscopes, which limits their progress and the quality and impact of their work. To acquire a modern 300 KV FEG liqHe microscope with associated accessories, space, and renovations, most PIs would have to orchestrate support from public, private, and institutional sources. The only successful acquisition to date through agency sources required simultaneous applications to NSF, the Keck Foundation, local foundations, and contributions from the department and various deans. Many PIs are superb scientists, but lack the knowledge, patience, political acumen, and time needed to orchestrate such a complex funding effort.
(11) Conclusion: The format of the review and funding systems do influence what research is done and how it is conducted. Because of the pressure to produce incremental progress on biology over the term of a 4-year funding cycle, it appears that important long-term technical development work has been neglected in favor of shorter-term, sometimes redundant, efforts to produce information of immediate biological significance.
The standard in-house microscope for routine work, specimen work-up, refinement of procedures, etc., is now a 200 kV with FEG. To be competitive, new investigators need daily access to an instrument of this caliber. A number of established investigators are using obsolete microscopes and need to upgrade.
For state of the art work, a 300 kV, FEG, liqHe instrument is essential. These should be shared by groups of investigators and operated continuously.
Anticipated computing demands will be three to four orders of magnitude greater than present, so significant investment in computing hardware will be necessary.
High-caliber, career-track support personnel are needed to maintain 300 KV microscopes in peak operating condition with round-the-clock availability to users.
As the sophistication of the instrumentation increases, more support personnel will be needed in individual laboratories.
With the transition to automation and high-throughput operation, more programmers will be needed for software maintenance and development.
Support for R&D projects targeted to hardware and software development is essential. Appropriate peer review arrangements should be made for R&D applications.
Funding opportunities should be created for investigators from the fields of physics, mathematics, materials science, engineering, and computer science for collaborative R&D projects (for example, "glue grants").
Outreach to scientists in other fields is required to recruit needed expertise. One recruiting approach is to organize workshops or courses to publicize research opportunities and invite established scientists and engineers from other fields.
Interdisciplinary graduate and postdoctoral training is needed.
Development of improved EM capabilities is critical for optimal progress in emerging integrative research in molecular cell biology, which depends on a structural approach in conjunction with genomics to defining the functions of molecular machines and networks.
Ten to 20 years of concentrated effort on technology development may be needed to realize the practical potential of EM in biology. Long before then, the underdevelopment of the technology will be an expensive bottleneck. Until the capability for direct visualization of macromolecular arrangements in very large assemblies and intact cells is developed, indirect approaches that are much slower and more expensive will have to be customized for each class of problems. This will delay the pace of progress in many important areas of integrative cell biology. While these capabilities are under development, transitional technologies, in particular cellular tomography, should be developed for high-throughput so they can be applied as routine structural tools.
Equipment and support. A top priority is acquisition of the next generation of electron microscopes and support for their operation, especially high-caliber personnel.
Technology development. Opportunities for support for technology development should be publicized. Although the participants who attended this workshop now have a much better picture of the opportunities for flexible support, the broad community does not. An announcement in the NIH Guide of interest in technology development to improve EM capabilities would stimulate research, help recruit senior collaborators from other fields, have a favorable impact on review, and facilitate negotiation for institutional cofunding and infrastructure support.
David Agard (University of California, San Francisco)
Pamela Bjorkman (California Institute of Technology)
Bridget Carragher (Beckman Institute, University of Illinois atUrbana-Champaign)
Wah Chiu, Chair (Baylor College of Medicine)
David Davies (National Institutes of Health)
David DeRosier (Brandeis University)
Ken Downing (University of California, Berkeley)
Joachim Frank (Wadsworth Center, N.Y. State Department of Health)
Bob Glaeser (University of California, Berkeley)
Steve Harrison, Chair (Harvard University)
Jim Hurley (National Institutes of Health)
Jennifer Lippincott-Schwartz (National Institutes of Health)
Paul Matsudaira (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology)
Dick McIntosh (University of Colorado)
Ron Milligan (The Scripps Research Institute)
Mary Porter (University of Minnesota)
Alasdair Steven (National Institutes of Health)
Susan Taylor (University of California, San Diego)
Nigel Unwin (Medical Research Council, Laboratory of Molecular Biology)
Please direct inquiries to:
James F. Deatherage, Ph.D.
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
NIH Building 45, Room 2AS.13J
Bethesda, MD 20892-6200
Tel: (301) 594-3828
Fax: (301) 480-2004
E-mail: deatherj@nigms.nih.gov
Ms. Teresa Trent, who made all of the administrative arrangements and ran the meeting.
Dr. James Cassatt, who provided support and guidance.
Dr. Jean Chin, who assisted with the running of the meeting.
Drs. Jean Chin, Catherine Lewis, Sue Shafer, and Don Schneider, who took notes.
