
National Institute of General Medical Sciences
Bethesda, Maryland
October 30-31, 2008
PART I. INTRODUCTION
Goals of the Meeting
The goal of the meeting, as outlined in the attached Meeting Charge, was to consider potential future directions for NIGMS funded structural genomics efforts. The meeting included a review of progress under the currently funded Protein Structure Initiative, but this was intended in the spirit of learning about what capabilities for high-throughput coordinated structural genomics have been achieved and could potentially be applied to new problems in the future. It was not intended as a repeat of the PSI Assessment, which was held in September, 2008. However, at the meeting a number of issues raised during the PSI Assessment clearly came to the fore, along with actions taken during the past year that begin to address them. Among the recurring themes were: i) evolution of the intellectual drivers for structural genomics; ii) engagement of a broad scientific community in the selection of protein structural targets; iii) improvements in experimental and computational functional annotation; iv) enhancing the utilization of the intellectual and material products of structural genomics through improved access, education, and dissemination.
Invited Speakers and Other Meeting Participants:
Speakers were invited to represent investigators from the currently funded protein structure initiative; members of the PSI Advisory Committee; members of the NIGMS Advisory Council; and a selection of scientists from the community at large who had no previous involvement with the PSI and no obviously preformed opinions regarding its merits. Representatives of other genomics, proteomics, and structural genomics initiatives, as well as of academia, government, and industry, were also included.
Materials Provided to the Meeting Participants:
Attached to this report are the Agenda, Structured Discussion Questions, Roster of Invited Speakers, and Other Participants. Also appended is the Detailed Table of Contents of meeting materials. These materials were distributed via a meeting-specific web site that included many useful links to other web sites as well as the meeting documents. The most important items were mailed to the participants on CD-ROM in advance of the meeting, provided in hard copy at the meeting, and emailed individually and personally to the meet the needs of invited speakers for information. The meeting materials are available at: http://fsgim.psi-structuralgenomics.org. Access is restricted at the moment, but the site will be made public once privileged communication manuscripts posted there have been published.
The Role of PSI Investigators' White Papers
Among the documents provided to meeting participants, were several white papers prepared by the currently supported investigators or based on PSI supported activities during the past year. The main white paper, "Opportunities for Structural Genomics Beyond 2010 – Creating Partnerships for the Future", provided progress reports for each of the centers, reports on biomedical theme projects, a consensus of the investigators views on opportunities for the future, and individual center views on the same topic. A second white paper dealt with the utility of models of varying quality. A third white paper dealt specifically with opportunities for structure determination by NMR. Additional papers were submitted by the investigators and others regarding coverage of sequence, family, fold space and the leverage of experimental results to additional protein sequences.
The meeting was not intended to explicitly review these PSI investigator white papers. However, the documents did get significant attention. Essentially ALL of the major ideas presented in these papers came up for discussion at the meeting and, in most cases, were endorsed as reasonable and, in some cases, as exciting ideas for future structural genomics initiatives. The meeting included a presentation by Andrej Sali that explicitly discussed recommendations of the white paper on modeling. Although no specific presentation or reference to the NMR white paper took place, the participants included several experts in NMR methods and the utility of this approach to future structure initiatives was well endorsed.
Capture of the Meeting Discussion and Recommendations
Speakers submitted biosketches and abstracts of presentations in advance of the meeting and their presentations were saved for use by NIGMS staff at the meeting. A science writer, Karin Jegalian, was tasked to capture as much of the meeting verbatim as possible and the text of this transcript was reviewed by NIGMS staff. Each of the meeting participants was asked to submit bullet points pointing to their views regarding future structural genomics initiatives. For various reasons, most chose not to provide these bullets in advance of the meeting, but DID provide individual recommendations after the meeting. These were shared with the meeting chairperson, David Eisenberg. Dr. Eisenberg crafted Part II Chairperson’s Summary. NIGMS Staff prepared Part III. Highlights of Background Documents and Meeting Presentations. Both have been circulated to the speakers for their comments and correction.
MEETING CHARGE
FUTURE STRUCTURAL GENOMICS INITIATIVES MEETING
October 30 & 31, 2008
Natcher Conference Center, Room D
Building 45, NIH Campus
Bethesda, MD
The goal of this meeting is to consider potential future directions for the NIGMS funded structural genomics efforts. The charge is not to determine whether or not to continue the Protein Structure Initiative (PSI). This decision will be made by the NIGMS staff with advice from the National Advisory General Medical Sciences Council (NAGMS Council) at a later date. Rather, the goal of this meeting is to develop and evaluate various potential alternatives for coordinated efforts taking advantage of the capabilities for high-throughput protein structure determination developed over the course of the PSI. It will include input from the broad scientific community as well as from those who have participated in the planning, execution, and analysis of the PSI to date. The objective is to suggest strategies that would enhance the value of the structural genomics and/or high throughput structural biology results to a broad scientific audience and ways to take best advantage of the resources that have been developed by the PSI. Discussion of the role of structural genomics and/or high throughput structural biology as distinct from other aspects of structural biology and the impact of structural genomics on other areas of science is within the scope of discussion for this meeting. Consideration of other areas of structural biology that may benefit from coordinated research effort is also relevant. The balance between the NIGMS investment in larger scale coordinated research as opposed to smaller scale investigator-initiated research should not be a focus of this meeting. This is a topic frequently discussed in a broader context by NIGMS staff and the NAGMS Council.
Key issues to be addressed include: i) prioritization in structural target selection, particularly the relative merits of structural coverage of protein families that have no or little structural coverage versus addressing targets of high biological interest; ii) integration of functional studies with structural studies; iii) the uses and limitations of homology models in driving biomedical research; iv) exploitation of intellectual and material products of the current structural genomics pipelines; v) involvement of the broad biomedical research community in all aspects of structural genomics efforts; vi) continued technology development for all aspects of protein structure determination and molecular modeling; vii) appropriately scaled structural genomics components and mechanisms of support under multiple budgetary assumptions.
FUTURE STRUCTURAL GENOMICS INITIATIVES MEETING REPORT
October 30 - 31, 2008
National Institute of General Medical Sciences
Bethesda, Maryland
PART II. CHAIRPERSON’S SUMMARY by Dr. David Eisenberg
A panel of ~30 scientists and scientific administrators identified and discussed the opportunities and needs in the area of Structural Genomics [SG]. The following is a brief summary of the outcomes of the discussion.
The panel was in uniform agreement that PSI-1 and PSI-2 have met many of the initial goals. In particular, new methods and technologies have been developed; high throughput structural pipelines have been set up in a number of centers; it has been demonstrated that macromolecular structures can be determined on the scale of many hundreds of structures per year, with both X-ray and NMR methods contributing; the cost per structure has fallen continuously; the range of previously unknown folds and protein families has been vastly enlarged; and several joint projects with biomedical communities have been established.
A. Scientific Problems. The panel judged the following scientific questions most ripe for applications of SG in a PSI-3:
- Possible further assessment of the number and nature of protein domain folds and protein families. This would continue the main application of SG in PSI-1 and PSI-2. Some computational scientists believe that up to 80% of protein sequence space can be defined by single domain that families have already been discovered, and hence, the protein universe consists of a finite set of single domain families. Various estimates suggest that around 25% of this space is already covered by representative structures. Completing the survey would have the reward of completeness, but could take much effort. Objections to continuing this goal as the main application of U.S. funded SG include the apparent lack of interest in this question in the biomedical community, and the apparent lack of relevance to immediate biomedical needs. Some note that domain structures fall far short of protein structures, which are the backbone of biology. In response to this point, it is important to note that the structures of targeted domains have in many instances been determined in the context of full proteins. Thus, domain interactions, as well as domain folds, have been determined. Also, as new folds have emerged from structures determined, knowledge of the extent of the protein universe has been extended. A more successful extension of this area in the future would ideally involve closer collaboration between a structural center and computational scientists. One such proposal is for consideration of structural coverage at the superfamily level as one component of U.S. funded SG. Future work should be built on an assessment of the full impact of the PSI in this area, including analyses of the past advances in molecular modeling, functional assignment, annotation, and structure determination by molecular replacement.
- Protein complexes: both predicted/overexpressed and isolated. This application would draw on the PSI technologies of expression, purification, crystallization, and structure determination. The results would help to illuminate biological networks and pathways. The complexes should include protein-protein, protein-RNA, and protein-DNA, as well as macromolecule-ligand complexes. This application would interface well with the genetics community. Further technological development is necessary.
- Eukaryotic proteins. The present PSI machinery is fully appropriate for eukaryotic proteins. Structure determinations will be larger projects and more expensive with slower throughput and output. A simple count of structures determined will not be an appropriate metric of achievement for structures of eukaryotic proteins (as well as for complexes).
- Membrane proteins make up nearly 30% of all proteins, but constitute less than 1% of known structures, and are particularly important for therapeutics. More advanced technologies must be developed.
- Microbiomes. Structures of proteins abundant in biological niches are appropriate for SG scale work.
- Protein and RNA structures from pathogens and model organisms are ideal candidates for SG.
- Functional determination of proteins. Both experimental and computational methods are needed. Amortizing proteins purified in a screen for common functions is a useful added step (as presently done in the joint Canadian-British- Swedish Structural Genomics Consortium).
B. Technology Development
Technology development was a highlight of PSI-1 and -2, and is ripe for extension at this time. Continued technology development is important for improving and optimizing all aspects of structural genomics methodology, including target selection, cloning methodologies, construct optimization and domain identification, purification, crystallization, NMR and X-ray data collection and structure determination, structural analysis, and homology modeling. Such development is best done under the umbrella of a PSI-type initiative. Technology proposals do not fare well in the regular study section system. Also, the final stages are often the most time consuming and expensive, and most important for dissemination of the technologies and ease of use by scientists other than the developers. Technology can be developed in large centers, similar in scale to the PSI-2 production centers, or in medium-sized centers, somewhat larger than the PSI-2 specialized centers. These are some areas of proposed focus:
- Membrane proteins: improved methods of expression, purification, and crystallization are required.
- Complexes: methods both for purified complexes and overexpressed/reconstituted complexes are needed. Bioinformatics will be important for advancing prediction of complexes. New screening methods are needed to validate complex formation.
- Related to complex formation, improved methods are needed for prediction of protein solubility and crystallizability.
- NMR is especially valuable for molecules that cannot be crystallized. Methods for larger molecules also need to be developed. NMR and crystallography approaches can be complementary to one another.
- Microcrystallography can vastly expand the range of structures that can be determined. The current needs include highly focused beams, and improved methods for crystal mounting, manipulation, centering, and robotic sample changing (on the scale of 500-1000 samples at a time).
- Modeling techniques, including homology model building, pseudo-atomic models built into EM densities, and model building from multiple data types at multiple scales. If protein modeling continues as part of PSI, computational scientists could be more tightly coupled to structural centers, so that new structures would help in improving potential functions, and bioinformatics insights would guide targeting of the specific interests of individual centers.
C. Outreach to the Biomedical Community
A major failing of PSI to date has been an effective outreach to both structural biologists and the non-structural biomedical community. Some suggestions to improve this situation:
- PSI centers need to focus on a post-analysis of structures that they produce, making the results accessible through means beyond mere PDB deposition. The PSI SG Knowledgebase will play an important role, but the centers need also to focus on the significance of structures they produce, so that their output is in accessible form and provides meaning and context to the discoveries. An annual review from PSI centers, illustrating and commenting on structures determined that year could help and provide a mechanism for assessing the actual value of the PSI structures and their use by the community.
- PSI centers need to translate their technological developments (e.g. robotics) into other labs. Workshops and visiting-scientists programs can help, but there must also be effective means for disseminating information on robotics, cloning, expression, crystallization, and so forth. Passive diffusion is not working well enough. It is noted that the PSI annual bottleneck meeting each Spring will be expanded this year (2009) to address ‘Enabling Technologies for Structural Biology’ to disseminate such information to the community. It is critical to continue support of the PSI SG Knowledgebase and the Materials Repository to aid in translation of the work of PSI to the wider biomedical and biological communities.
- More frequent talks at National, International and Society meetings by SG scientists, particularly outside of the usual Structural Biology and Structural Genomics meetings, such as ACA, IUCr, ISGO, Keystone Symposia, Gordon Conferences, etc.
- Outreach to the biomedical community on applications of SG to various fields of biomedicine.
- Avoidance of competition with projects in non-PSI structural laboratories to the extent possible.
D. Mechanisms for support of PSI activities and interfacing with the wider biomedical community.
This was the most difficult area of discussion, on which there was least consensus of the panel. There WAS agreement that the PSI centers need to focus on problems of biomedical significance, without competing with RO1 and PO1 research. But how can this be achieved?
All believe that it is important to maintain and further develop high-throughput structure determination centers. Technology development can take place in these, as well as in mid-size centers or small centers. Some believe that properly selected biomedical projects can thrive in such centers, but others suggested that the centers need to abandon the lead for projects with a specific biomedical focus. In PSI-2, ~15% of the total effort is applied to such biomedical theme projects chosen by each of the large scale production centers, 15% to community targets and 70% to the communal PSI Network targets. All agree that proposals for a specific biomedical focus need in the future to be subjected to the community review process. Some believe that biomedical projects are best developed in small and medium centers, which would be judged on the quality of proposed research. Nor can the large centers become purely service centers. They need some flexibility for development of their own work, and the ability to collaborate with each other as well as with small and medium-sized centers and with RO1s and PO1s. In PSI-2, the centers, worked well together as a unified team under the auspices of the U54 mechanism. Target selection, the setting of objectives, metrics, milestones and production goals, and interaction between production and specialized centers, have been under the direction of the Operation Management Group (OMG), the Bioinformatics groups (BIG4) , and the PSI Steering Committee. The ability to coalesce and unite as a team has substantially enhanced the success and the productivity of the centers, both individually and collectively.
Submitted by Dr. David Eisenberg, with suggestions from Ian Wilson, Dorothee Kern, Wesley Sundquist, Frank Raushel, and John Norvell.
Part III. HIGHLIGHTS FROM BACKGROUND DOCUMENTS AND MEETING PRESENTATIONS
DAY 1
A. Basic Facts About the Protein Structure Initiative:
Jeremy Berg and John Norvell provided background information on the PSI and its role within the NIGMS Strategic Plan.
The Protein Structure Initiative currently supports four large scale centers (ca $10 million each); six specialized centers (ca $3.5 M each); two homology modeling centers ($1.3 M total) and a series of R01s focused on improving modeling ($1.9 M total); the PSI-Materials Repository ($1.0 M); the PSI-Structural Genomics Knowledgebase ($2.6 M); and a small supplemental grants program. The total budget for FY2008 was $68.1 million total costs. This budget supports a total of 97 investigators with independent PI status at their institutions: Large Scale Centers (30 PIs); Specialized Centers (50 PIs); Modeling Centers (10 PIs); Materials Repository (3 PIs); Knowledgebase (4 PIs).
Putting this in perspective, the NIGMS research grant portfolio is organized into about 60 program areas, each of which cover a problem of approximately the same scientific scope as the PSI. The typical NIGMS research grant portfolio includes from 50 to 250 awards with an average budget of $343,000/award = total of $17.1 – $85.7 million total costs/program area. (NIGMS Research Portfolio Data – FY2004). Thus, the PSI is approximately equivalent in size (both budget and number of investigators supported) to any other of the NIGMS portfolios of grants. The primary difference is that instead of being a collection of individual R01 awards, each separately solicited, reviewed, and awarded; the PSI is a coordinated network of investigators and grant support that was solicited, reviewed, and awarded in an intentionally structured series of competitions. The PSI, although a large initiative, represents only 3.5% of the overall NIGMS budget of $1.9 billion.
Administration of the PSI differs from that of other research grant portfolios in being highly coordinated through a series of steering committees and outside advisory committees. The projects have been awarded as cooperative agreements and have involved extensive NIGMS staff participation and oversight. A series of regular annual meetings, workshops, and regular teleconferences have facilitated information exchange among the groups. A hallmark of the PSI has been the degree of cooperation and coordination among the investigators in attacking an over-arching common problem.
Lila Gierasch, representing the PSI Advisory Committee, explained the role of the committee, and its involvement with the program and with NIGMS staff. She noted that the PSI is an experiment in big science and noted both the productivity of the centers and the stunning effectiveness of group target selection. Chris Sander, another member of the PSI Advisory Committee, reinforced her comments complimenting the investigators on their ability to transcend the individual competitiveness of research and to form a network of exceptional added value. He noted the administrative overhead of investigators, advisors, and NIGMS staff that is needed to make it work, but thought this a useful way to organize science to encourage collaboration.
B. Accomplishments of the Protein Structure Initiative
Ian Wilson – representing the large scale centers; Wayne Hendrickson chair of the PSI Steering Committee and representing the small scale centers; Helen Berman – representing the Knowledgebase; and Joshua LaBaer – representing the Materials Repository gave reports on the progress of the current PSI and ideas for the future. Their comments are included within the overall report below, rather than in the presented order.
From a technology development and implementation standpoint, it is clear that the PSI has been very successful in establishing high-throughput pipelines for protein structure determination for some classes of proteins. This has involved both the automation and the optimization of many steps in the process. These activities have provided a general benefit to the structural biology community.
The long-range goal of the Protein Structure Initiative (PSI) is to make the three-dimensional atomic-level structures of most proteins easily obtainable from knowledge of their corresponding DNA sequences (From Mission Statement). PSI-1 (FY2000-2005) tested whether such a goal was feasible by developing new methods and technologies, constructing structure determination pipelines, and establishing new policies for openness regarding work in progress and immediate access to the results. A total of 1,383 structures were solved. PSI-2 (FY2005-2010) has continued the development of technology while implementing the production phase of high-throughput structure determination. A total of 1,871 (as of date) have been solved in PSI-2 (grand total = 3,254 and expected to exceed 4,000 by the end of PSI-2). For comparison, the total number of non-redundant protein structures in the PDB is around 20,050 (at 95% identity level as of 12/1/08). The average cost per structure has dropped from around $670,000 to $57,000. In order to optimally explore new sequence/structure and fold space, the PSI has concentrated on proteins of low sequence identity. Seventy-four percent of the PSI structures were in the =30% identity range. In contrast, 72% of all other structures in the PDB, are for proteins with high (=90%) sequence identity.

Figure: Comparison of Sequence Redundancy of PSI with other PDB Structures. Data table.
Analysis included in a pre-publication manuscript from Burkhart Rost shows that structural genomics is the largest contributor of novel protein structures. During the past two years, the PSI contributed just over 10% of all structures deposited into the PSI, but over 35% of all novel structures.
Beyond the raw numbers of structures solved, there is considerable controversy about the goal of structural coverage, leverage of solved structures, and the value of models for proteins at varying levels of sequence identity. The PSI Assessment released in December, 2007, raised some doubts about progress toward the goal in the mission statement. Although fold space appeared to be nearing complete coverage, sequence space is continuing to grow linearly, making the PSI an open ended endeavor. Coverage of protein families at a level of 30% sequence identity was thought to be unbounded and it was concluded that the PSI was only making modest headway. The analysis cited in the PSI assessment relied on work by Michael Levitt. He has since conducted a different analysis that was included as a pre-publication manuscript in the background materials to the October, 2008 meeting. This analysis focused on single domain families and concluded that the growth of single domain families is saturating at about 25,000 domains. The continued growth of sequences comes primarily from multi-domain families. Structures are known for a quarter of the single domain families and half of all sequences can be partially modeled from those structures. An important point, of course, is that multi-domain proteins can be modeled by assembling single domain protein models. Further, it should be noted that most PSI protein structures are of full-length proteins (typically 200-500 residues in length), not isolated protein domains.
According to data presented by John Moult, the number of families of a given size follows a power law. There are many small families (>20,000 with only one member) and many fewer large families. Families including three or more members make up 87.7% of sequence space. While the number of singleton families continues to grow, the number families with 5 or more members appears to be saturating at around 9,000 and even the number of doubleton families is leveling off. Additional analysis in the Rost paper indicates that by systematically targeting large families of previously unknown structure, the leverage of PSI structures is over four-fold higher than that for non-PSI structural biology efforts. Thus, by judicious choice of further targets, the PSI should be able to achieve its goal for a large fraction of the sequence universe.
For the most popular model organisms, structures or structural models at >30% sequence identity can be computed for from 28% to 51% of their sequences. For humans, the coverage is 35%. If one accepts a different level of model accuracy, as reflected by Gene3D or Genthreader, then models can be constructed for 75% to 90% of sequences. By focusing structural genomics on a more restricted universe of sequences and structures, it should be possible to reach 100% coverage for these key organisms at the superfamily level.
More analysis of the data is needed, but the meeting participants were reasonably enthusiastic about structural coverage at the superfamily level as one of the goals of future structural genomics initiatives.
Ian Wilson reported progress on the initial 1269 Pfam families of unknown structure that had been assigned to the large scale centers three years ago in October, 2005. One or more representative structures have now been solved for 33% (22% covered by the PSI, 11% by others including primarily the Japanese and European SG centers). He pointed out the PSI is the only group focusing on proteins of unknown structure and function. PSI centers have solved 275 novel structures of domain families of unknown function. Reevaluation of proteins families of unknown function in light of new data suggest that 1700 remain.
Wayne Hendrickson provided a brief overview of the entire PSI and its management via the PSI-2 Steering Committee and subcommittees. He also provided a brief sketch of the activities of each of the six specialized centers. These are described in detail in the PSI Investigators’ White Paper. In general, these specialized centers have focused on technology development and/or challenging classes of proteins, notably eukaryotic proteins and membrane proteins from any source. Advances have been made particularly in areas of protein expression, labeling for NMR, crystallization, identification of protein complexes, and chaperone assisted crystallography. A significant percentage of the PSI total of eukaryotic and human protein structures have been solved by specialized centers. Considerable progress has been made toward establishing HT-approaches for membrane proteins and a number of solved structures have resulted.
Helen Berman provided an update on activities of the PSI-Structural Genomics Knowledgebase (KB). Functions of the KB include capture of all information on PSI activities, including: target selection (TargetDB); progress, materials, and methods (PepcDB); experimental structures; models derived from protein structures (Protein Model Portal); annotation of structure/function; publications. It is the major tool for outreach to the broader scientific community, including nomination of targets, user search of PSI information, access to the Materials Repository, and of course linkage to the PDB structure repository. A partnership with Nature Publishing Group has established a Nature Gateway that regularly provides editorial features on the latest structural and technological advances, related news and events, and a research library of PSI publications and articles of broader interest. Although, this effort was only recently launched, the PSI SGKB has already addressed many of the issues raised by the PSI Assessment.
Joshua LaBaer provided an update on the PSI Materials Repository (MR) which is operated by the Harvard Medical School Institute of Proteomics (PlasmID repository). Since 2006, a total of 9,933 plasmids, including empty vectors, have been processed by the MR. Initial lags due to preparation at both ends of the process have delayed submissions, but these are expected to reach steady state quickly with a total of over 100,000 PSI plasmid to be archived. About 40,000 are ready for deposit and 30,000 have been submitted. Thus far, 21 user orders have been processed for 31 PSI generated plasmids. This is expected to increase quickly as the resource becomes populated and the PSI collection becomes better known. For comparison, PlasmID contains over 97,000 plasmids and distributed about 13,000 plasmids in response to over 400 requests in past three quarters. The process for archiving deposited samples features many quality control steps, including barcoding, recloning and full-length sequencing, annotation, and redundant automated glycerol stock and freezer storage systems. Curated plasmid information is entered in the PlasmID database and linked to the PSI-KB. Negotiations with all donor sites and many potential users has resulted in expedited materials transfer agreements. Materials are distributed at cost to users with a PSI discount this is currently $30 per clone; $1200 per 96-well plate. Information on the PSI specific plasmid collection is available through the PSI-MR Portal, which links to the PDB and the PSI-KB modules. This is part of an aggressive outreach strategy to make material products of the PSI more widely known.
C. Utility of Models to the Broad Scientific Community is the Key to the Success of any Future Structural Genomics Initiatives
As noted above, the selection of targets, the density of coverage required, and the leverage of solved structures are all dependent on modeling capabilities. The ultimate goal is also not simply to produce models, but to produce models that are actually used by researchers to advance the nation’s research agenda.
A key variable emphasized by each of the speakers Andrej Sali, Catherine Peishoff, and John Moult is that the required accuracy of a model depends on the purpose for which it is used. Whereas highly accurate models may be needed for drug design or to inform detailed mechanisms of action, lower quality models can provide insight into a variety of problems. In the absence of something better, researchers will and should use whatever models are available if they satisfy minimal accuracy criteria. Therefore, it is important that users are aware of the limitations of the models. Because of the need for accountability in modeling, experts in modeling need to be involved in research that relies heavily on results of models. Examples where homology models are useful included:
- Annotation of approximate function, functional domains, alternative splicing, epitope selection (low accuracy)
- Mapping of interaction sites and locations of SNPs to assist interpretation of molecular mechanisms of disease (medium accuracy)
- Development of ortholog sets and evaluation of SNP impact (high accuracy)
- Detailed function and drug design (very high accuracy)
- Integrative (hyrbrid) methods for structure determination (x-ray, NMR, EM, MS) and application to solution of higher order structures, complexes, metabolic pathways, large assemblies, in combination with experimental data (all accuracies).
Andrej Sali provided a report on the Workshop on Applications of Protein Models in Biomedical Research held by the PSI on July 13-14, 2008. Among the recommendations of the workshop were development of standards for publication of models, formats for data and software exchange, standards for assessment of models, and outreach to a broad scientific community to provide awareness of the strengths and limitations of models. A Protein Model Portal (Torsten Schwede, PI) is under-development as part of the PSI-SG Knowledgebase that will facilitate these activities and either serve or potentially compute models upon user request. Thus far, the state of the art in modeling does not appear to have been much impacted by the PSI, although the field has made use of the many new structures. For example the PSI has been a major contributor of blind datasets for the CASP (Critical Assessment of Techniques for Protein Structure Prediction) meeting. However, it is early and new methods are under development. The best mechanisms for engaging modelers and experimentalists in any future structural genomics initiatives was discussed at length. Three potential mechanisms were suggested: i) stand alone modeling; ii) modeling as an integral part of PSI center activities; iii) modeling outside the PSI based on user needs and traditional collaborative arrangements
Sali presented a figure that nicely illustrates the relationship between model accuracy and its potential application.
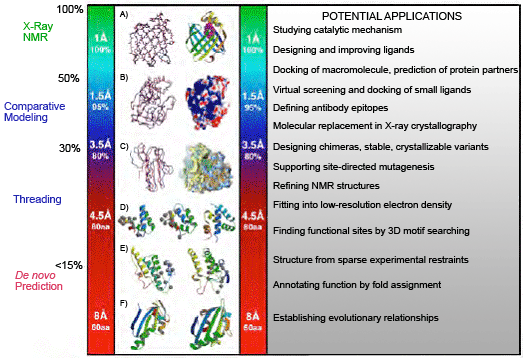
The figure "relationship between model accuracy and its potential application" illustrates the degree of structural information detail available as a function of the nominal resolution of the data over a range from 1 Angstrom to 8 Angstrom resolution. At 1 Angstrom, individual atoms may be seen. At 8 Angstrom only the overall folded pattern of the protein chain can be resolved. The figure includes potential applications of data at each resolution ranging from studying catalytic mechanism which requires very high resolution data to establishing evolutionary relationships which can be accomplished with low resolution structural data.
Catherine Peishoff presented results of an informal survey among pharmaceutical, biotech, and specialty chemical companies regarding the use of homology models. Most do use such models and in most cases constructed these in-house with some limited usage of published models from other groups. Models were used for a variety of purposes relating to both structural biology efforts and compound design. Whereas improvements to methods and models would be welcomed, modeling without reference to specific applications and users was not considered valuable. Incorporation of additional information beyond sequences, such as associated ligands, would be interesting. An increased emphasis, both experimentally and computationally, on membrane-bound proteins (including many druggable targets) would be welcomed. Publication of coordinates and supporting data for all published models should be required. A long discussion about the merits of virtual screening suggested that this should NOT be undertaken by future structural genomics centers, unless suitable expertise is included (but see Frank Raushel comments below). The recently funded NIGMS Drug Docking and Screening Resource at the University of Michigan (Heather Carlson, PI) was mentioned as a possible partner for such activities. Despite the lack of immediate and obvious benefits of the PSI structures to the pharmaceutical industry, Dr. Peishoff thought there had been tremendous infrastructure benefits from the PSI and other structural genomics initiatives, and that they should be continued.
As part of PSI-2, NIGMS funded two modeling centers to develop and improve homology modeling techniques. In addition, seven research grants (R01s) focused in this area were funded as part of PSI-2. These efforts started just two years ago. The impact is not yet clear.
D. Opportunities for Future Structural Genomics Initiatives and High-Throughput Structural Biology
1. Coverage of Sequence/Structure Space could be continued and as discussed above can be brought to some satisfying level of closure with additional effort. This requires rethinking the extent of the sequence universe for which total coverage (or leverage) is desired, the accuracy of model that is acceptable, the specific targets that are needed. Ian Wilson presented a figure from the PSI Investigators’ White Paper that nicely illustrates the concept of focused structural genomics, where the capabilities of high-throughput pipelines could be brought to bear on a subset of sequence space determined by particular biological problems of interest.
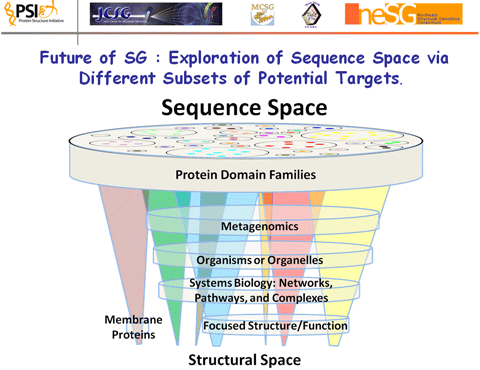
The figure "Exploration of Sequence Space via Different Subsets of Potential Targets" illustrates the entire universe of sequence space as disk. Subsets of this universe shown as cones extending downward from this disk represent structure space, i.e., particular subsets of sequence space for which structures have been solved or can be adequately modeled. Additional disks sequence/structure space represent regions of particular interest which may be examined in greater structural depth than is possible for the entire universe. Examples shown include the topics described immediately below the figure.
Megafamilies. Understanding how structural diversity translates into functional diversity is one such potential driving biological problem. This idea would stress the solution of additional structures for very large families, where even though one or more family representatives have been solved, additional structures are needed to be able to model all family members at high accuracy. Examples already being investigated include the FMN binding split barrel family.
Metafamilies. These are families of proteins that are over-represented in genomes from selected environments, such as the human gut microbiome, and that contain many novel proteins of unknown function. Recent sequencing efforts have changed the view of several protein families that were previously thought to be small or rare in nature.
Complete Structural Genomes for Specific Organisms. Significant progress has been made toward the complete coverage of the structural genome for the simple model organism Thermatoga maritima. Similarly focused efforts could be pursued for other widely used model organisms, protein families known to occur in humans, or human pathogens. Structural efforts should tie into other "omics" work in these organisms.
Systems Biology. Complete structural coverage for all proteins in regulatory networks, metabolic pathways, or complexes could be a driving biological problem. Numerous such efforts are underway at the genomics, transcriptomics, proteomics or other informational levels. Structural efforts would both benefit from and contribute to other efforts in this area.
Leveraging results by weighting both the contribution of a given target to the goal of covering sequence protein/folding space and the value of a given structure to a driving biological problem would provide for a win-win outcome.
2. Metagenomics. The study of genomes from communities of organisms that cohabit a particular environment may be a fruitful area for structural genomics. Claire Fraser-Liggett gave an update of research on the human gut microbiome, which has well established relevance to human health and disease states. Research is revealing the differing densities and species of microorganisms through different parts of the GI tract and the inter-individual variability in microbial populations. The NIH Roadmap Human Microbiome project already supports research on genomics, metabolomics, proteomics, GC profiling, cloning and sequencing, and RFLP mapping. 16S rRNAs are being used to determine the numbers of species, their abundances, their genetic relationships, and correlation with disease. Key biological problems include the bidirectional signaling mechanisms between microbes in the gut and between microbes and the host. How do gut microbes interact with the endocrine and immune systems? High-throughput sequencing centers for metagenomics have just been established with a goal of establishing reference genomes for around 1,000 organisms (cultured and non-cultured). It is anticipated that their activities will lead to recognition of many new proteins some of known function, but many of unknown function. Growth of formerly small protein families into megafamilies is anticipated. Many new targets for structure determination, some suitable targets for drug development are expected.
3. Functional Structural Genomics. A criticism of the PSI has been that it is focused primarily on determining structures and not contributed to understanding the function of many of the proteins studied. This is because of the conditions under which it was agreed with the NIGMS Advisory Council that the PSI would operate when it was established; i.e., that the PSI would focus on structural coverage as a resource for the broad biological community and that it would not compete with the broad community in the area of protein function. There is clearly enthusiasm among both the investigators and the general community for improving functional characterization. Frank Raushel discussed work in collaboration with Brian Shoichet and John Gerlt that has been directed to establishing the function of proteins of unknown function that have been solved by the PSI. The approach focuses on superfamilies of generally known enzyme reaction class (e.g. amidohydrolase), then uses the computational docking of potential ligands (e.g., from the KEGG database of ~12,000 metabolites) to solved structures and homology models, along with information about operon structure to make hypotheses about possible substrates. These are tested by screening libraries of compounds, then followed up by detailed enzymological studies. Examples included the identification of a Thermatoga protein (Tm0936) as an S-adenosyl-L-homosysteine and 5 methyl-thioadenosine deaminase. Further work in the enolase and amidohydrolase superfamilies has led to new functional annotations for 32 previously known reactions.
Jacquelyn Fetrow suggested functional annotation as a driving force for PSI target selection. PSI centers could collaborate with investigators interested in molecular function to better understand sequence/structure/function relationships. She mentioned the weakness of common computational approaches to function annotation based primarily on sequence homology and the valued added by structure alignment. However, she referred to the multiple levels at which annotation may be directed and the multiple functions that a protein may have that are not necessarily conserved. She noted that methods that predict functions at the top levels of the GO function hierarchy are not sufficiently detailed to be useful to experimentalists. Annotation needs to link local sequence/structure to functional sites, mechanisms, substrate/inhibitor specificity, and to link the specific function with biological processes and networks. She provided examples from work on the peroxidoxin family. She also discussed three examples of proteins which had been assigned as serine hydrolases by chemical genomics methods (i.e., activity probe for nucleophilic serine) that could be identified based on local folding motifs as an entirely different class with dehydrogenase activities.
The best way to implement an approach to functional structural genomics is not clear. Some of the current centers are capable of doing HT-screening as well as HT-structural biology. However, the value of screening, unless well focused was questioned. NIH also already supports a number of HT-screening operations and collaboration with those centers might be more efficient. Similarly, virtual screening may be better carried out in collaboration with experts (e.g., Brian Shoichet), rather than adding further to the demands of the structural centers. Further functional studies might operate through separately funded partnerships. In later discussion, it was noted that there is a backlog of 1,500 structures of unknown function in the PDB. It was also noted that HT-functional studies could be pursued using PSI proteins in the absence of solved structures. These are key questions for any future structural genomics initiatives.
4. Biomedically Important Targets. Three speakers were invited to address biomedically important targets as drivers for structural genomics efforts. Cheryl Arrowsmith described the Structural Genomics Consortium cofunded by the Canadian and Swedish governments, three pharmaceutical companies, and several foundations. The overall budget for this multi-site consortium ($21.79 M USD in 2007-2008) is about one-third that of the PSI. Funders have nominated ~3,000 targets, which are primarily human or human parasite proteins (i.e., all eukaryotic) and mostly of known general functional class. Since 2004, ~600 human structures and ~80 parasite structures have been solved. These have been significant contributions to the total number of known human (10%), malarial (90%), and known drug targets (20%) in the PDB, although in many cases other members of the protein family have been solved. About 1/3 of the proteins were associated with a ligand. Most of the SGC structures have been derived from recombinant proteins expressed in E. coli. Almost all were structures of protein domains, not necessarily complete multi-domain, multi-chain proteins. However, all structures were functional in that they bind to their cognate ligands and/or have the correct enzymatic activity. Within families of proteins, comparisons of active sites, substrate/inhibitor binding profiles, and other functional studies have been carried out to identify substrates for orphan family members and identify novel inhibitors. Example biological problems driving the work have included analysis of protein kinase families relevant to cancer and enzymes of chromatin modification in epigenetics. The infrastructure of the SGC is similar to the PSI in that tools and resources have been developed to follow progress on targets, a repository exists for useful biological materials, and the SGC has invested effort and resources to make their results and technologies readily available to others. However, the SGC is not as high-throughput as the large scale PSI centers, as might be expected for eukaryotic targets. It also differs from the PSI in pursuing only human targets and not considering homologous proteins from other species (except in the parasitology program which targets orthologs from 7 human parasites). Arrowsmith argued for the value of structural genomics as a tool for both discovery and explanatory science. Each approach may be more cost effective for certain problems and a balance should be sought between the two. She emphasized the value of the proteins produced and of conducting systematic, but limited functional characterization of the proteins (primarily focused on identification of molecules which interact with a given protein) in parallel with structural studies. She suggested a number of possible biomedical themes generally similar to those enumerated by Ian Wilson above.
Wayne Anderson discussed activities of the NIAID funded centers for structural genomics of infectious diseases, which is one component of the larger suite of NIAID “omics” programs. Each center is a multi-site effort. The structural centers are operated under contract with targets assigned from a list of specifically targeted organisms. Target selections emphasizes biomedical relevance and potential therapeutic benefits and are not necessarily unique or novel structures. Priorities include known drug targets or their orthologs, essential genes, virulence factors, pathogenic factors, markers of infection, vaccine candidates, drug resistance proteins, and taxonomically-specific enzymes. The majority of the ~2,700 targets currently selected are from infectious bacteria. The centers seek community input and seek to complement community research efforts. Requests from outside the center are evaluated for feasibility and approved by NIAID before work begins. The goal is to determine ~400 structures over a 5 year period. In addition to structural work, the centers do some chemical screening. Complexes with a bound ligand are a priority for both stabilizing the protein, to gain insight into biological function, and as a starting point for drug design. Both x-ray and NMR methods are used for fragment-based lead identification. Anderson acknowledged contributions of the PSI to technology used by the NIAID centers and also felt the efforts were complementary. Among ideas for future structural genomics initiatives, he favored some continued coverage of the protein structure universe, metagenomics – particularly comparisons of pathogen and commensals, organism focused genomics, biomedically important targets, and networks and pathways.
Timothy Veenstra was asked to discuss some of the cancer focused "omics" efforts, including the Cancer Genome Atlas program. These are in general oriented toward finding biomarkers of cancer and of therapeutic response. Thus far, there is not a structural component to these genomic efforts. Veenstra focused particularly on proteomics efforts based primarily on mass spectroscopy. He cautioned that these efforts are generating too many biomarkers that have not been adequately validated. A problem may be that the most relevant pathological samples are not being examined. Other problems relate to sample preparation and data integration. Correlation between biomarker results from genomic, transcriptomic, and proteomic efforts have not been consistent. It is difficult to know which if any targets should be a high priority for structure determination. A more hypothesis driven approach may be needed. Ways that these efforts could interact with the PSI, include target selection, protein expression and production, and identification of protein ligands, domains, and protein-protein interactions.
5. Challenging Problems – Systems, Complexes, Networks, and Membrane Proteins
Three speakers were asked to address these topics and the technology development that would be needed to address them in the future. David Eisenberg discussed the significance of protein complexes and advances in methods to predict protein-protein interaction. Study of complexes may allow determining the structures of proteins that are predicted to be disordered in the absence of binding partners, better understanding of protein function in vivo, and elucidate roles of proteins in networks and pathways. He discussed the ProLinks database of pathways and complexes inferred from genomic analysis. He also referred to bioPIXIE (Pathway Inference from eXperimental Interaction Evidence). From a technology development standpoint, he highlighted microcrystallography for its ability to examine smaller crystals and thus extend the range of proteins that can be studied. Advantages include, more homogenous crystal lattices, better diffraction, ability to work with proteins that tend to form microcrystals. Work carried out using beamlines at ESRF in France allowed work on amyloid crystals down to 1.5 micron in size. Such capabilities are currently under development for the Advanced Proton Source (APS) in this country. In addition, improvements are needed in all aspects of crystal handling, mounting, centering, sample changing. With respect to target selection, he proposed Gretsky's principle (i.e., skate to where the puck will be, not where it is now). Much of structural biology supported through other methods is focused on structures of proteins that are known to be important. The interactions of these proteins are often ignored in the short run. By working on proteins that are suspected to interact with proteins currently under study, structural genomics efforts can avoid direct conflict with individual investigators to provide structural information that will become important in the future as a fuller picture evolves. He provided an example of predicted protein interactions involved in RNA splicing. NIGMS staff note that a good many projects are focused on determining the structures of single proteins or domains of proteins that are part of extremely large complexes. Would it be useful to approach this type of problem in a high-throughput parallelized approach, rather than incrementally, one protein at a time?
In a much later discussion during the meeting, Steven McKnight brought up the potential of studying isolated intact native complexes, rather than single recombinant proteins. Looking at complexes that have been made by the cell, should satisfy concerns about biological relevance.
Wayne Hendrickson addressed the difficulty of studying membrane proteins and eukaryotic proteins. Membrane proteins are predicted to be 25-30% of all proteins, yet are only <1% of structures in the PDB. As of October 1, 2008, there were 405 solved structures (174 unique proteins) of which 30 (24 unique proteins) had been solved by NMR. The first structure of a membrane protein appeared 25 years after the first soluble protein and though the rate of solution of both soluble proteins and membrane proteins is increasing exponentially, the rate of growth for membrane structures is slower. From discussion it was clear that the PSI large scale centers and other structural genomics centers have mostly avoided membrane proteins. Two specialized centers of the PSI have focused exclusively on membrane proteins and others have contributed some work to new methods development. Challenges include bioinformatics without large structure database, undetected obligatory partnerships, complications from detergent solubilization, instability outside membrane environment, poor crystallization and slow optimization, and NMR dampened by detergent micelles. Among membrane proteins, eukaryotic proteins have been a particular problem, except those that occur in naturally rich sources such as mitochondrial or retinal membranes. However, some recent successes from the PSI specialized centers include the aquaglycerolporin from P. falciparum, human AA2 adenosine (caffeine) receptor, X4 transporter, and TrkG. Opportunities for future structural genomics projects focused on membrane proteins include a large number of receptors, pumps, channels, and enzymes of great biomedical importance. Methods under development for many years (e.g., solid state NMR, NMR with oriented bilayers, and solution NMR in detergent micelles) are finally delivering structures. Methods for expression and crystallization, including as antibody complexes and fusion proteins, are becoming established. Pipelines for membrane proteins and genomic scale experiments in expression, solubilization, and crystallization are possible.
In discussion there was generally strong support for continued structural genomics efforts on membrane proteins. It was noted that eukaryotic proteins in general should also become a more significant focus.
Scott Lesley was asked to discuss technology development issues in general. He noted that membrane proteins, protein complexes, eukaryotic proteins in general are challenging targets and that these have mostly not been pursued by the large scale HT centers to date. With respect to membrane proteins, improvements are needed in protein expression, solubilization, characterization, and stabilization of protein conformation including cross-links, fusions, or antibody complexes; crystallization methods, including cubic and other lipid phases, methods to detect crystals; and microbeam synchrotron sources to collect data from smaller crystals. For complexes, a challenge is determining which proteins form complexes. Methods in use include 2-hybrid screening, TAP-tag/MS methods, isolation of native complexes from cells, and bioinformatics predictions. Each method has its strengths and weaknesses. Some success has been achieved in the co-expression of proteins that form complexes, but additional innovations and reductions in cost and production time are needed. Eukaryotic proteins in general remain a challenge and non-E. coli based systems, including yeast, baculovirus, mammalian cell cultures, and in vitro expression have been explored with some success. These need to be scaled up to enable high-throughput structure determination. He suggested some challenging problems that are being pursued. These include complete target sets, determinants of molecular recognition, and prediction of function from structures. He mentioned the need for new screening methods to predict function, methods for identifying protein-protein interaction, methods for annotating proteins of unknown function. Complete coverage for pathways, networks, or specific organisms, requires salvage methods for proteins that do not yield to the initial pipeline approach. He suggested the inclusion of additional structural methods combining cryoEM, as well as modeling constrained by NMR data, in addition to x-ray crystallography.
E. Community Engagement – Targets, Service, Dissemination
Not unexpectedly, this proved to be the most challenging session of the meeting. Five scientists we asked to represent the interests of the structural biology and cell biology communities at large. Mario Amzel discussed results of an informal survey among colleagues at Johns Hopkins and elsewhere. All agree that structural information can be important. They relied on publications to learn about new structures. Some had searched the PDB for coordinates, but none had specifically searched for or used results from the PSI. They were unconvinced that the structure of a protein with only 30% identity to their own proteins of interest would be useful and had little interest in validating models for their protein, rather than working toward solving the actual structure. Those surveyed did not think the PSI should continue studies aimed at covering sequence/structure space, unidentified proteins, and rare folds. Future work should be driven by biologically important problems and should engage other investigators working in the chosen areas. The key problem is deciding which structures to do. Normally this is a part of peer review. All those interviewed indicated that PSI centers working on problems driven by the biological questions should not be funded as a Center but should compete through the standard R01 study sections with other non-PSI investigators working on different or related problems. Amzel also presented some interesting data on structure publications in the highest profile journals – Science, Nature, Cell and noted a decline in recent years of work from the U.S. In the area of membrane proteins, he noted that European, Japanese, and HHMI funded groups continue to dominate. Whether this trend reflects on the PSI or some other aspect of U.S. funded research is unclear.
Dorothee Kern emphasized the problem of ignorance on both sides of the discussion. The community is generally unaware of what is going on in the PSI and many are not sufficiently familiar with the potential value of the structural models to their work. The PSI-KB needs to be successful, but in addition, the PSI centers need to work much, much harder on outreach. Beyond basic structures, important problems will require understanding protein dynamics and its role in their function. Modeling can contribute here to examine high energy states that are not in the crystal structures. For example, ligand binding to a protein that is in dynamic equilibrium may occur. Minor states may be the key to activity.
Barry Gumbiner stated that he came as a cell biologist and developmental biologist who has collaborated with structural biologists, and without any preconceived notions of the PSI. He thought it important to make the program more widely known. It is important to advertize the PSI-KB and to help it stand out. There are already many gateways on the Nature site. This will not be enough. He thought homology models can be very powerful tools, when used properly with acknowledgement of the level of accuracy. He mentioned that there are hundreds of models of cadherin, but it is difficult to know which are good models. Atomic level models give the illusion of atomic level resolutions, which they may not possess. A useful role of the KB would be to help educate non-experts on how models should be used. With regard to coverage of protein space versus solving structures with biological or medical relevance, he would favor the latter. This means working on proteins of known function and may mean overlap with research already on-going in other labs. Community involvement will be greater if targets include proteins of greater interest. At this point infrastructure developed by PSI-1,2 should be turned to more complex problems.
Michael Snyder emphasized the importance of improved communication. The broad scientific community does not know about the PSI and he thought the consortium was not marketing itself as aggressively as other similar sized efforts. The PSI has surprisingly little visibility at other “omics” meetings. If the program had better outreach, perhaps more people would use its products; however, many of the targets are esoteric. The goal of “solving all structures” was a good goal and it is somewhat disappointing that it still seems out of reach. At this point there is a need to pick more selected structures and these might as well start with proteins that people are interested in studying. Workshops to spread technology would be worthwhile. Future directions should include more complex proteins and may need to include additional methods such as cryo-EM. Structures of RNA and protein-RNA complexes could be added. Functional annotation of structures is important and could benefit from assay automation. Future structural genomics efforts should complement and partner with other large scale biology efforts. Lots of reagents, clones, proteins, and antibodies are generated by other projects that could benefit structural genomics and vice versa. With respect to conflicts with R01 funded science, this may be unavoidable, but should not inhibit the application of high-throughput approaches if these are truly the most useful.
Wesley Sundquist indicated that as a structural biologist himself, he was impressed by the level of throughput achieved by the PSI, and doubted that many people fully appreciated this accomplishment. He was reasonably supportive of the goal of structural coverage, since this appears to be working and is what HT-centers do well. It also does not conflict with work on R01s, but does not yield results that most scientists care about. Unless modeling improves, there will be a preference among scientists to solve the structure that they are interested in, rather than relying on a model. For the future, an increased emphasis on eukaryotic proteins and disease-related proteins would be appropriate, and a decrease in throughput and increased cost per structure would have to be acceptable. However, both the results and the intermediate reagents would be more useful. He would favor a mixed portfolio of targets from easy through complex, including community nominated targets. A means of soliciting community nominations was just put in place. Whether the HT-centers can adapt themselves to many disparate targets, as well as the ultimate success of the PSI-KB and PSI-MR remain to be seen. Collaborating with other consortia may be an effective way to engage a broader community with many targets that people will care about.
In the general discussion, the following points came out. 1) The value of models may be taken as a given by structural biologists, but not so much by others who are more closely aligned with a particular organism and biological problem. Additional education and outreach is needed. Several examples were discussed where structures solved by the PSI provided useful models and insights. 2) The number of community targets is still low and centers are accommodating the requests. 3) A difficulty with community targets is the impedance mismatch between the HT centers which process 96 samples at a time in a few days versus collaborators who typically have a few proteins ready at a time. 4) Pipelines are currently efficient for soluble prokaryotic proteins. Additional technology development will be needed to reach anything similar for eukaryotic proteins, membrane proteins, and complexes. 5) The rewards system for investigators in structural genomics has been tied to the number of structures solved. Some weighting needs to be applied to the difficulty and impact of the structure. Advisory committees should consider these factors in setting goals and milestones for future initiatives and may wish to consider more tangible ways of rewarding progress (e.g., through administrative supplements). 6) The utility of structures of unknown function and the utility of functional characterization, now or in the future, as part of structural genomics or as an activity outside the structural consortium was discussed at length. A hypothesis of the PSI has been that structures will be found useful in time and that widely-conserved proteins are especially likely to be important. These may be valid hypotheses. However, there are also many proteins of known function without known structure for which solving the structures should be a higher priority.
DAY 2
Morning Session I. Readback and Additional Discussion
Four individual were asked to serve as recorders for discussions during Day 1 and they were asked to read back their summaries. Jacquelyn Fetrow summarized the discussion on the current status of the PSI. The existing program has demonstrated that high-throughput structure determination can be accomplished. Difficult problems/challenges for the future include multi-protein complexes, membrane proteins, and eukaryotic proteins in general. Submission of community nominated targets through a single coordinating portal is a good development. The PSI-Structural Genomics Knowedgebase and Materials Repository are perceived as good, but additional effort is needed to make them well known. The recurring discussions through Day 1 about the utility of models to whom, for what purposes, at what levels of accuracy was recapped. A broader survey of this issue may be useful. Which aspects of model development, virtual screening, and functional annotation should be taken up by the PSI centers is up for debate. Some such work is already in progress. Data on screening is being deposited in PubChem.
Mario Amzel summarized the discussions about sequence coverage and metagenomics. This summary recapitulated the issues about trade-offs in coverage versus acceptable quality of model. Further discussion did not resolve the issue of whether there is a need to continue expansion of the database. Discussion of metagenomics repeated the idea that the PSI could contribute to hypothesis driven research in this area and could take the lead. However, the issue of funding through a center rather than through competing R01s came up again. Ian Wilson noted that several thousand targets have already been assigned. The relationships between metagenomics and mega-families was further clarified. The basis for target prioritization was again discussed. The idea of pursuing thermophilic eukaryotes was mentioned.
Dorothee Kern summarized points from the biomedical targets and challenging proteins sessions. There was consensus on the importance of functional annotation. A small number of successful examples were presented. Docking works for cases with small conformational changes on binding or where structures of several conformations can be modeled. Another consensus was the importance of experimentally determining function and the need to do so on a large scale. The impact of structures from the SGC is felt to be high. It is surprising that almost all of the targets have been satisfactorily expressed in E. coli. Both the SGC and the NIAID centers have had closer interaction with biology than the PSI centers. The PSI directors would welcome the opportunity to be more involved with functional studies. Various cancer biomarker projects could be useful partners for PSI, but care is needed that the targets are well validated. There is general consensus on the importance of membrane protein and complexes, signaling networks and metabolic pathways as future targets, and on the need for continued technology development within the PSI. In discussion, the level of activity needed or feasible in functional follow-up was discussed. The current supplements for function project, while a nice thing, is very modest. A scheme for prioritizing functional studies will be needed. R01 or P01 grants to make use of the PSI structures could be prioritized through peer review. The combined use of cryo-EM and HT-crystallography of subunits, e.g., for reconstruction of the nuclear pore, was suggested as an area for technology development.
Scott Lesley summarized the discussion on community engagement. Structure is considered important, but there may be a general lack of knowledge about the PSI and how to utilize structures that it is producing. The Knowledgebase and Materials Repository are useful, but there are many additional things that they can do. There is some question about the utility of homology models to the general community. Target selection based on biological relevance versus contribution to expanding protein structure space was discussed and there is enthusiasm for continuing this effort for a more focused universe, including human targets. In discussion, there was considerable enthusiasm for competing coverage for key model organisms, especially yeast, worm, and mouse. How competitive review could be used to select targets was discussed and the cost/benefit to whom of overlap with R01 projects was considered.
David Eisenberg asked Ian Wilson to review high points from the PSI Investigators’ White Paper to be sure that all these topics had surfaced. Wilson noted the following:
1) Coverage of the protein universe is an open-ended and complex question, but could be finished to some intermediate level of satisfaction. With 1,500 folds currently known, the completion of fold space would require about 400 more folds to be solved.
2) Metagenomics is a useful driver for structural targets. In discussion, it was not clear what contribution the structures will make to answering biological questions about the human microbiome in the near term, but this can be refined.
3) Coverage of individual organisms would be feasible. About ½ of all Thermatoga protein families have been completed as a demonstration project. Work on other prokaryotes and eukaryotes is feasible.
4) Systems, networks, signaling pathways, and complexes would provide challenging work for future structural genomics initiatives and lead to enhanced interaction with the other communities of science.
5) Focused structure/function studies for given families would better understanding protein function and evolution.
6) Membrane proteins are clearly a priority for the future genomics initiatives.
7) Collaborations with outside groups and consortia would link the structural pipelines to biological problems.
In discussion, differences between the idealized pathways in textbooks and in real pathways in specific organisms were noted. In addition to metabolic pathways (substrate-product relationships), there is a need to think about how proteins are organized physically, including other types of pathways, such as signaling pathways, redox pathways.
The desirability of finishing off sequence space at some level of resolution, perhaps near 100% coverage at the threading level, was suggested. Beyond solving more structures, it is important that the lessons learned be boiled down to a reasonable number of basic principles that can be articulated well in a few reviews and understood by all. Participants were considerably impressed by the progress that has been made to date and there was enthusiasm for continuing effort. Preferably, each structure solved can provide a win-win result by both addressing an important biological problem and also contributing to our understanding of protein sequence/structure/folding space.
Morning Session 2. Program Prioritization and Structure
The chairperson, David Eisenberg, suggested constructing a number of menus of options that NIGMS should consider:
1) Large biological problems that could be solved by structural genomics.
2) Different mechanisms for forming consortia to solve these large scale problems.
3) Achieve outreach to the rest of the biomedical community.
4) Related technology development.
Going through the list of topics suggested by the PSI Investigators’ White Paper and by others, there were few that were rejected as inappropriate. Some were viewed as more likely to succeed. The discussion turned more on how the work in these subject areas could be parsed to utilize the strengths of HT structural genomics, including ability to use an entire network or centers, rather than just a single center.
With regard to functional studies, once the centers have produced proteins, they should take advantage of this accomplishment. Screening may be accomplished through collaboration with other individuals or centers in appropriate collaborations or partnerships. Some activity might be carried out in the structural centers, but there is concern about diffusion of effort.
Concerning technology development, there are many, many areas for further improvement. Inclusion of cryo-EM as an HT center activity is doubtful. However, this may be useful in a collaborative fashion. Several centers include collaborations in this area already. Development of microcrystallography, on the other hand, is exactly the sort of thing the centers should be doing. Tomography and computational methods for solution of large structures might also be useful as collaborative activities. NMR has proven to be useful in the PSI for proteins of limited size. Technology development would enable larger proteins to be studied. In a very general sense, technology development to solve biologically driven problems should be a centerpiece of future structural genomics centers. Furthermore technology translation and dissemination should be one of their metrics of success.
Whether modeling should be part of the program, integral to all centers, or funded through stand alone centers, or through individual R01s, was discussed without clear conclusion. The choice depends on the goal. If fold space as a goal is dropped, then modeling research could be substantially curtailed. Some modeling is currently done within the existing structural centers. A role for computation and modeling groups in the future is to help interpret what has been accomplished. Much time is spent on target selection, but little on reporting the results beyond depositing the structures.
The subject of community outreach and partnerships spurred lengthy discussion of the need for additional analysis of the output of the PSI and mechanisms to disseminate results. Participation in meetings, BLOGs, visiting scientist programs, workshops, were suggested. A marketing plan and a responsible person to take the lead is essential. Other products for dissemination include standard operating procedures. Integrating information activities with other information centers, PubMed and Genebank, was noted, as was the need to make material available more graphically.
The key issue of how to pursue biological problems in collaboration with outside groups and without engendering competition with and resentment by individual structural biology labs was discussed but not resolved. Community nominations may help. Working on targets that individual labs would not or could not otherwise do may also help. However, excessive avoidance of other peoples “turf” may mean continuing to have lower impact than is desirable. An option is to make resources available, but people running the centers would not want to simply provide service. However, there is nothing that says the centers cannot put their own biological drivers on the table for discussion. Self-assembled groups, working on a specific area, topic, or organism might form a partnership with a center. These way, the center investigators would not be competing with the R01 community, they would be part of the community. How to review these collaborations is a question, but it is important that this activity be transparent.
Another issue in community outreach and partnerships is the handling of joint information. The PSI has operated with different rules regarding intellectual property. Targets have been declared in advance, progress on targets reported at given milestones, and structures posted immediately upon completion. These policies, particularly release of coordinates upon deposition to the PDB, will likely be continued. Will they apply to collaborators as well as to structural genomics centers?
The intended closing discussion about structure and scale of future structural genomics initiatives was truncated for lack of time. Large scale centers are needed if a high-throughput capability is to be maintained. Obviously, they need to be openly competed. If the activities of the centers are broadened beyond simply structure determination, then those activities need to be competed. How to separate review of the capabilities of the centers from the biological driving themes (and whether this is a good idea), remain to be determined. It is unlikely that any of these activities can fit within a typical R01 application and the CSR standing study sections for review. An advantage to the large scale U54 mechanisms is that it allowed new things to be taken on when unanticipated opportunities appeared. A minimal size of center is needed to accomplish anything in the way of technology development. Biological collaborations could conceivable range from R01s to P01s and this is already working in PSI-2.
Concluding Remarks
At least two major models were proposed during or after the meeting for organization of any Future Structural Genomics Initiatives:
Model A. This would follow roughly the model of the current PSI with modifications. It would include at least 3, possibly 4, large scale centers that would conduct high-throughput structure determinations. It would include 2, possibly more, specialized centers focused on technology development for membrane proteins and complexes. In Model A, the driving biological problems could be jointly shared among the network of centers, and these could be determined in advance (as part of the RFA and/or by the applicants through peer review), or elaborated in a separate RFA to establish collaborations.
Model B. This model would significantly alter the landscape by reducing the differences between large scale and specialized centers. Each center would have its own intrinsic driving biological problems that would be judged along with their capabilities in a single RFA. The centers could have overlapping interests and operate within a network or could operate substantially independently.
The PSI-Structural Genomics Knowledgebase and the PSI-Materials Repository would continue and extend their present functions in either model. The roll of the current Homology Modeling Centers remains to be determined.
Many questions remain to be addressed for the organization under either scenario. Many possibilities lie in between Model A and Model B. These will be taken up by the PSI Advisory Committee at its annual meeting in December, 2008, and by the National Advisory General Medical Sciences Council at its meeting in January, 2009. Based on these discussions, it is anticipated that NIGMS staff will develop Concept Clearances for consideration of the Council. Assuming Council does approve of a forward-going structural genomics initiative, then RFAs will be issued in late spring, early summer of 2009 for fall 2009 submission and funding in FY2010.
Report prepared by Peter C. Preusch, NIGMS with the assistance of the speakers and NIGMS staff, particularly, Ward Smith, Charles Edmonds, John Norvell, and Catherine Lewis.
AGENDA
Future Structural Genomics Initiatives Meeting
Wednesday, October 29, 2008
Evening Pre-Meeting Reception - Local Restaurant or Hotel
7:00 p.m. – 9:00 p.m.
Informal gathering for those who are in town and pre-meeting discussion with chair and session chairs to clarify assignments.
Thursday, October 30, 2008 Natcher Conference Center, Room D
I. Introduction 8:30 – 9:00 a.m.
Introductions, Meeting Agenda, and Mechanics – Peter Preusch
Meeting Charge and Rationale – Jeremy Berg
Introductory Comments by Meeting Chair – David Eisenberg
Overview of PSI Administration – John Norvell
II. Current Status of the PSI – Presentations by representatives of PSI-2
9:00 – 10:30 a.m.
Speaker 1 – PSI Large Centers – Ian Wilson
Speaker 2 – PSI Steering Committee and Small Centers – Wayne Hendrickson
Speaker 3 – PSI Knowledgebase – Helen Berman
Speaker 4 – PSI Materials Repository – Joshua LaBaer
Speaker 5 – PSI Homology Modeling – Andrej Sali
Coffee Break 10:30 a.m. – 11:00 a.m.
Speaker 6 – Industrial Use of Models – Catherine Peishoff
Speaker 7 – PSI Advisory Committee – Lila Gierasch
III. Future Directions in Structural Genomics Discussions
Topics and Keynote Speakers
11:30 a.m. – 12:30 p.m.
Topic 1: Structural Coverage
Keynote: John Moult
Topic 2: Metagenomics
Keynote: Claire Fraser-Liggett
Lunch Break 12:30 – 1:30 p.m.
Thursday, October 30, 2008 Natcher Conference Center, Room D
III. Future Directions in Structural Genomics Discussions (Continued)
Topics and Keynote Speakers (Continued)
1:30 – 3:00 p.m.
Topic 3: Functional Structural Genomics
Keynote: Frank Raushel, Jacquelyn Fetrow
Topic 4: Biomedically Important Targets
Keynote: Cheryl Arrowsmith, Wayne Anderson, Timothy Veenstra
Topic 5: Challenging Problems – Systems, Complexes, Membrane Proteins
Keynote: David Eisenberg, Wayne Hendrickson, Scott Lesley
Coffee Break 3:00 – 3:30
Topic 6. Community Engagement – Targets, Service, Dissemination
3:30 – 6:30 p.m.
Keynotes: Mario Amzel, Dorothy Kern, Barry Gumbiner, Pamela Silver, Michael Snyder, Wesley Sundquist
Evening Reception and Dinner – Local Restaurant (TBD)
6:30 p.m. – 9:00 p.m.
Discussion Session Topic Chairs Write-up Discussions Overnight
Friday, October 31, Natcher Conference Center, Room D
V. Morning Session 1. Readback and Additional Discussion
8:30 a.m. – 10:00 a.m.
Readback of discussion summaries from Day 1.
General discussion of topics and other issues arising.
Coffee Break 10:00 a.m. – 10:30 a.m.
VI. Morning Session 2. Program Prioritization and Structure
10:30 a.m. – 12:00 a.m.
Considerations for organization and scale of future activities.
Meeting Adjourned by Noon.
STRUCTURED DISCUSSION/STUDY QUESTIONS
Future Structural Genomics Initiatives Meeting
(Addressed in part during speaker presentations)
Session I. Basic Information about the PSI
- What have been the mechanisms of support and management of the PSI?
- How much has been invested in the PSI relative to other areas of NIH investment?
- What were the conclusions of the PSI assessment and what actions have been taken since the assessment to address concerns?
Speakers: Jeremy Berg and John Norvell
References:
Basic Administrative Information – Tab 8; RFAs - Tab 9.A
PSI Assessment Report, Intro, and Commentary – Tab 9.B, C, D
Session II. Current Status and Future of the PSI
Large Scale Centers, Specialized Centers, PSI Materials Repository, and PSI Structural Genomics Knowledgebase
- What have been the accomplishments of the PSI to date?
- What are the technological capabilities of the PSI centers?
- What types of materials and information have been produced and are they being utilized by the community at large?
- What opportunities exist for utilizing the infrastructure developed by the PSI centers?
Speakers: Ian Wilson, Wayne Hendrickson, Joshua LaBaer, Helen Berman
References:
PSI Investigators’ White Paper – Tab 7 – Progress Reports
PSI Meeting Reports and Advisory Group Summaries – Tab 9.E,F,G,H,I
Session II. Current Status of the PSI (Continued)
Homology Modeling and the Use of Models
- For what types of problems are current homology models of sufficient accuracy for a given level of sequence identity?
- Are the developers of new methods for homology modeling making use of the output of the PSI? If yes, how dependent are they on this source of new structures? If no, why are they not making better use of the PSI structures?
- How much improvement in modeling can be expected during the next few years and what are the primary obstacles to improvement?
- How can models and modeling expertise be made more readily available to non-expert users?
Speakers: Andrej Sali and Catherine Peishoff.
References:
a) PSI Investigators’ White Paper – Tab 7 – Section by Burhard Rost and Adam Godzik.
b) Report of Workshop on Applications of Protein Models in Biomedical Research by Torsten Schwede and Andrej Sali – Tab 9.R.
Advisory Committees and Operations Management of the PSI
- Has progress of the PSI generally been satisfactory?
- What activities and structures of the PSI have worked well? What has not worked well?
- What has been the level of PSI oversight and is it the right level?
- Has the level of interaction with NIGMS staff been adequate and appropriate?
- What recommendations would you make for the organization of future structural genomics initiatives?
Speaker: Lila Gierasch – PSI Advisory Committee Member
References:
Basic Administrative Information – Tab 8.
PSI Advisory Committee Meeting and Update – Tab 9.F
Session III. Future Directions in Structural Genomics Discussions
Topic 1. Structural Genomics – Progress, Done, Doable?
- What size is sequence space and how is it changing? What is the size of protein structure space and how is it changing? What are the relationships between sequence space, structure space, protein fold space, and protein family space?
- What is the current status with respect to coverage of protein structure space? How many families of what sizes exist for given definitions of a family? For how many of these are example structures known? For how many domains? For how many protein structures? What priority should be given to continuing the original goals of the PSI-2?
- Would a more focused approach be more or less useful and how long will it take to achieve various milestones for structural coverage?
- What percentages of which genomes are currently covered by known structures for given levels of sequence identity and model accuracy? This should be addressed for the human genome, the top ten model organisms, and other selected organisms or systems of interest, such as key pathogens or specific organelles. What opportunities exist for future activities?
Speaker: John Moult
References:
PSI Investigators' White Paper – Tab 7.
Novel Leverage of Structural Genomics – Tab 9.Q
PSI Statistics update provided by Burkhard Rost – Tab 9.Q
The Nature of the Protein Universe by Michael Levitt – Tab 9.T
Topic 2. Metagenomics
- Will metagenomics contribute more to growth in the number of novel single domain families or more to growth in the size of families of already known domains?
- How much is metagenomics expected to contribute to the growth of novel single domain families compared to other sources of sequence information?
- How valuable will structures from metagemomic sequencing efforts be if the species of origin is unknown?
- Will fragments of proteins be of value if the entire coding gene is not know?
- What priority should be given to human commensual organism metagenomics versus environmental metagenomics?
Speaker: Claire Fraser-Liggett
References:
PSI Investigators’ White Paper – Tab 7.
PSI Statement on the Knowledgebase Website – Tab 9.L
Other Metagenomic References – Tab 9.L
Topic 3. Functional Structural Genomics
- NIGMS supported structural genomics efforts to date have focused primarily on the determination of structures. What can be done to enhance the connection between these structures and their functions?
- What experimental and theoretical approaches can be taken to assign functions to proteins of unknown function that utilize the solved protein structure?
- What can be done to improve the utilization of data from structural genomic programs by the research community at large?
- What important questions about protein structure and function could be further facilitated by structural genomics initiatives?
Speakers: Frank Raushel, Jacquelyn Fetrow
References:
Hermann, et al. – Tab 9.C – Articles cited in Assessment Report
A Protein Structure (or Function?) Initiative – John Gerlt - Tab 9.V - Commentaries
Topic 4. Biomedically Important Targets
- What prioritization schemes have been used to select biomedically important targets to date? What are the unaddressed needs and opportunities for future biomedical target emphasis?
- What contributions can specifically biomedically selected targets for structural genomics projects make to the overall problem of understanding protein structure and function?
- How can partnerships between structural genomics centers and other researchers contribute to the solution of such structures?
- What has been the value of the biological themes of the current PSI Large Scale Centers?
- What are the most compelling biological themes for the future?
Speakers: Cheryl Arrowsmith, Wayne Anderson, TBN
References:
A Future for the Protein Structure Initiative – Edwards – Tab 9.V
Center for structural Genomics of Infectious Diseases – Tab 9.W – Other Useful Links
The Structural Genomics Consortium – Tab 9.W – Other Useful Links
The Cancer Genome Atlas – Tab 9.W – Other Useful Links
Topic 5. Challenging Problems – Systems, Complexes, Membrane Proteins
- What are the opportunities and obstacles to the application of a structural genomics approach to more complex systems?
- Are there advantages to all solving structures for a given metabolic pathway or signaling network or organelle?
- What progress has been made in the methods and setting up pipelines for determination of membrane protein structures and are these proteins now amenable to high-throughput structural approaches?
- Is the technology currently in place adaptable to the determination of these challenging structures? If not, what future technology developments are needed?
- Would an organized research network like the PSI be more efficient than more traditional approaches to these challenging problems?
Speakers: David Eisenberg, Wayne Hendrickson, Scott Lesley
References:
PSI Investigators’ White Paper – Tab 7.
NIH Structural Biology Roadmap – Tab 9.W– Other Useful Links
Topic 6. Community Engagement – Targets, Service, Dissemination, Partnerships
- How can the biomedical research community broadly participate in the selection and prioritization of targets for high-throughput structure determination?
- How can the community make more effective use of the structures that are determined through structural genomics initiatives?
- What mechanisms exist for collaboration and/or partnerships between structural genomics research centers and individual investigators or groups of investigators (both structural biologists and non-structural biologists)?
- What other opportunities exist for coordinated structural biology research that can take advantage of the resources and infrastructure that have been created through the PSI.
Speakers: Mario Amzel, Dorothee Kern, Barry Gumbiner, Pamela Silver, Michael Snyder, Wesley Sundquist.
References:
PSI Investigators’ White Paper – Tab 7.
PSI Knowledgebase Summary – Tab 9.H
PSI Materials Repository – Tab 9.I
PSI/Nature Knowledge Index – Nature Gateway – Tab 9.J
PSI Framework for Community Nominated Targets – Tab 9.M
Session V. Readback and Additional Discussion
- Have the previous discussions adequately captured the important issues, pros, cons, and options for the topics that were introduced?
- Are there other ideas for future structural genomics initiatives that have not been covered in the preceding sessions?
- What relative emphasis should be given to various potential goals of any future structural genomics initiatives?
Session VI. Program Prioritization and Structure
- Are the resources of PSI-2 optimally deployed for future work?
- What components are essential for any future organized structural genomics activity? Are these activities appropriately supported and run well?
- What components could be equally well supported through investigator-initiated, peer reviewed proposals or other mechanisms?
- What organizational and oversight structures are needed? Are the existing structures appropriate or should they be changed? If so, how?
- What is the appropriate scale of activity, given the current budget climate? How would this differ under various TBN budget scenarios that may develop in the coming years?
References:
Basic Administrative Information – Tab 8.
Reports of Annual Meeting and Advisory Committees – Tab 9.E, F, G, H, I
PSI Investigators' White Paper – Tab 7.
PSI Calendar of Meetings and Events – Tab 9.X
Roster of Invited Speakers Future Structural Genomics Initiatives Meeting
Chairperson
David Eisenberg, Ph.D.
HHMI Investigator
Professor
Department of Chemistry and Biochemistry
University of California-Los Angeles
611 Charles Young Dr. East
Los Angeles, CA 90095
Tel: 310-825-3754
E-mail: david@mbi.ucla.edu
http://www.doe-mbi.ucla.edu/People/Eisenberg/
L. Mario Amzel, Ph.D.
Professor and Director
Department of Biophysics and Biophysical Chemistry
Johns Hopkins University
School of Medicine
725 N. Wolfe Street WBSB615
Baltimore, MD 21205
Tel: 410-955-3955
Fax: 410-955-0637
E-mail: mamzel@jhmi.edu
http://biophysics.med.jhmi.edu/amzelm/amzel.html
Wayne Anderson, Ph.D.
Professor
Department of Biochemistry, Molecular Biology and Cell Biology
Northwestern University
Feinberg School of Medicine
303 E. Chicago Avenue
Chicago, IL 60611
Tel: 312-503-1697
Fax: 312-503-5349
E-mail: wf-anderson@northwestern.edu
http://csgid.org/csgid/investigators.php
Cheryl Arrowsmith, Ph.D.
Chief Scientist
Structural Genomics Consortium
Ontario Cancer Institute
University of Toronto
MaRS South Tower, Room 705
101 College Street
Toronto, ON M5G 1L7
Canada
Tel: 416-946-0881
E-mail: carrow@uhnres.utoronto.ca
http://nmr.uhnres.utoronto.ca/arrowsmith/
http://www.TheSGConline.org
Helen Berman, Ph.D.
Professor
Department of Chemistry and Chemical Biology
Rutgers University
Wright Lab 610 Taylor Road
Piscataway, NJ 08854
Tel: 732-445-4667
E-mail: Berman@rcsb.rutgers.edu
http://rutchem.rutgers.edu/content_dynamic/faculty/helen_m_berman.shtml
Jacquelyn S. Fetrow, Ph.D.
Reynolds Professor of Computational Biophysics
Department of Physics and Department of Computer Science
Wake Forest University
301B Olin Physical Laboratory
P.O. Box 7507
Winston Salem, NC 27109
Tel: 336-758-4957
Fax: 336-758-6142
E-mail: fetrowjs@wfu.edu
http://users.wfu.edu/fetrowjs
Claire Fraser-Liggett, Ph.D.
Professor of Microbiology and Immunology
Director, Institute for Genome Sciences
University of Maryland School of Medicine
20 Penn Street
Baltimore, MD 21201
Tel: 410-706-3879
E-mail: cmfraser@som.umaryland.edu
http://medschool.umaryland.edu/FACULTYRESEARCHPROFILE/viewprofile.aspx?id=20004
Lila M. Gierasch, Ph.D.
Distinguished Professor
Department of Biochemistry and Molecular Biology and Department of Chemistry
University of Massachusetts
1224 LGRT
710 North Pleasant Street
Amherst, MA 1003
Tel: 413-545-6094
E-mail: gierasch@biochem.umass.edu
http://www.chem.umass.edu/Faculty/gierasch.htm
http://www.biochem.umass.edu/gierasch/index.html
Barry Gumbiner, Ph.D.
Professor and Chair
Department of Cell Biology
University of Virginia
P.O. Box 800732
Charlottesville, VA 22908
Tel: 434-243-9290
Fax: 434-924-2794
E-mail: gumbiner@virginia.edu
http://www.faculty.virginia.edu/gumbiner
Wayne A. Hendrickson, Ph.D.
HHMI Investigator
University Professor
Department of Biochemistry and
Molecular Biophyisics
Columbia University
College of Physicians and Surgeons
650 West 168th Street
New York, NY 10032
Tel: 212-305-3456
E-mail: wayne@convex.hhmi.columbia.edu
http://www.hhmi.org/research/investigators/hendrickson.html
Dorothee Kern, Ph.D.
HHMI Investigator
Department of Biochemistry
Brandeis University
MS 009
P.O. Box 549110
Waltham, MA 02454-9110
Tel: 781-736-2354
E-mail: dkern@brandeis.edu
http://www.bio.brandeis.edu/faculty01/kern.html
Joshua LaBaer, M.D., Ph.D.
Director
Harvard Institute of Proteomics
Harvard University
320 Charles Street
Cambridge, MA 02141
Tel: 617-324-0827
Fax: 617-324-0824
E-mail: josh@hms.harvard.edu
http://www.hip.harvard.edu/
Scott Lesley, Ph.D.
Director of Protein Sciences
Genomics Institute of the Novartis Research Foundation
10675 John Jay Hopkins Drive
San Diego, CA 92121
Tel: 858-812-1551
E-mail: slesley@gnf.org
http://www.gnf.org
Steven McKnight, Ph.D.
Distinguished Chair in Basic Biomedical Research
Department of Biochemistry
University of Texas Southwestern Medical Center
5323 Harry Hines Blvd.
Dallas, TX 75390-9004
Tel: 214-648-3342
E-mail: steven.mcknight@utsouthwestern.edu
http://www.utsouthwestern.edu
http://www.mcknightlab.com
John Moult, D.Phil.
Director and Professor
Center for Advanced Research in Biotechnology
University of Maryland Biotechnology Institute
9600 Gudelsky Drive
Rockville, MD 20850
Tel: 240-314-6241
E-mail: moult@umbi.umd.edu
http://moult.umbi.umd.edu
W. James Nelson, Ph.D.
Rudy J. and Daphne Donogue Professor
Professor of Biology and of Molecular and Cellular Physiology
Department of Biology
Stanford University
The James H. Clark Center, E200B
318 Campus Drive
Stanford, CA 94305-5430
Tel: 650-725-7596
Fax: 650-724-4927
E-mail: wjnelson@stanford.edu
http://nelsonlab.stanford.edu
Catherine E. Peishoff, Ph.D.
Vice President
Computational & Structural Chemistry
GlaxoSmithKline
1250 S. Collegville Road
UP12-210
Collegeville, PA 19426
Tel: 610-917-6584
Fax: 610-917-7393
E-mail: catherine.e.peishoff@gsk.com
Frank M. Raushel, Ph.D.
Professor of Chemistry and of Biochemistry and Biophysics
Department of Chemistry
Texas A&M University
College Station, TX 77842
Tel: 979-845-3373
E-mail: raushel@mail.chem.tamu.edu
Andrej Sali, Ph.D.
Professor and Vice Chair
Departments of Biopharmaceutical Sciences and of Pharmaceutical Chemistry
California Institute for Quantitative Biosciences
University of California-San Francisco
Mission Bay, Byers Hall
1700 4th Street, Suite 503B
San Francisco, CA 94158-2330
Tel: 415-514-4227
E-mail: sali@salilab.org
http://salilab.org/
Michael Snyder, Ph.D.
Lewis B. Cullman Professor
Department of Molecular, Cellular, and Developmental Biology
Yale University
Director, Yale Center for Genomics and Proteomics
KBT 926
P.O. Box 208103
New Haven, CT 06520-8103
Tel: 203-432-6139
E-mail: michael.snyder@yale.edu
http://www.yale.edu/snyder/
Timothy D. Veenstra, Ph.D.
Director
Laboratory of Proteomics and Analytical Technologies
National Cancer Institute
SAIC-Frederick
Fort Detrick, Bldg. 469/160
Frederick, MD 21702
Tel: 301-846-7286
E-mail: veenstrat@mail.nih.gov
http://web.ncifcrf.gov/atp/default.asp?LabID=11&page=labs
Wesley I. Sundquist, Ph.D.
Professor
Department of Biochemistry
University of Utah
Bioscience Graduate Studies
15 N. Medical Dr. East, Room 4100
Salt Lake City, UT 84112
Tel: 801-585-5402
E-mail: wes@biochem.utah.edu
http://www.bioscience.utah.edu/mb/mbFaculty/sundquist/sundquist.html
Ian Wilson, D. Phil.
Professor
Department of Molecular Biology
The Scripps Research Institute
10550 North Torrey Pines Rd.
BCC206
La Jolla, CA 92037-1000
Tel: 858-784-9706
E-mail: wilson@scripps.edu
http://www.scripps.edu/mb/wilson
NIGMS Staff
http://www.nih.gov/nigms/
Jeremy M. Berg, Ph.D.
Director
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2AN.12
Bethesda, MD 20892-6200
Tel: 301-594-2172
Fax: 301-402-0156
E-mail: bergj@mail.nih.gov
John C. Norvell, Ph.D.
Chief, Structural Genomics and Proteomics Technology Branch
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200
Bethesda, MD 20892-6200
Tel: 301-594-3832
E-mail: Norvellj@mail.nih.gov
Meeting Contact
Peter C. Preusch, Ph.D.
Chief, Biophysics Branch
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200
Bethesda, MD 20892-6200
Tel: 301-594-1158
E-mail: preuschp@mail.nih.gov
Other Meeting Participants - Future Structural Genomics Initiatives Meeting
Ravi Basavappa, Ph.D.
Program Director
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.19e
Bethesda, MD 20892-6200
Tel: 301-594-0828
Fax: 301-480-2004
E-mail: basavapr@nigms.nih.gov
Stephen Bryant, Ph.D.
Senior Investigator
Computational Biology Branch
National Center for Biotechnology Information
National Library of Medicine
National Institutes of Health
Room 5S504, Bldg. 38A, 8600 Rockville Pike
Bethesda, MD 20894, USA
Tel: 301-435-7792
E-mail: bryant@ncbi.nlm.nih.gov
George Chacko, Ph.D.
Chief, Bioengineering Sciences and Technologies IRG
Center for Scientific Review, National Institutes of Health
Bethesda, MD 20892
Tel: 301-435-1245
Fax: 301-480-4184
E-mail: chackoge@csr.nih.gov
Jean Chin, Ph.D.
Program Director
Cell Biology and Biophysics Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.19A
Bethesda, MD 20892-6200
Tel: 301-594-2485
Fax: 301-480-2004
E-mail: chinj@nigms.nih.gov
Charles G. Edmonds, Ph.D.
Program Director
Cell Biology and Biophysics Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.13K
Bethesda, MD 20892-6200
Tel: 301-594-4428
Fax: 301-480-2004
E-mail: edmondsc@nigms.nih.gov
Paula F. Flicker, Ph.D.
Program Director
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2AS.13H
Bethesda, MD 20892-6200
Tel: 301-594-0828
Fax: 301-480-2004
Fred K. Friedman, Ph.D.
Division of Biomedical Technology
National Center for Research Resources
National Institutes of Health
6701 Democracy Boulevard, Room 972
Bethesda, MD 20892-4874
Tel: 301-435-0775
Fax: 301-480-3659
E-mail: fred.friedman@nih.hhs.gov
Ann A. Hagan, Ph.D.
Associate Director for Extramural Activities
National Institute of General Medical Sciences, DHHS
National Institutes of Health
45 Center Drive, MSC 6200, Room 2AN.24H
Bethesda, MD 20892-6200
Tel: 301-594-4499
Fax: 301-480-1852
E-mail: hagana@nigms.nih.gov
C. Craig Hyde, Ph.D.
Scientific Review Administrator
Office of Scientific Review
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 3AN.18a
Bethesda, MD 20892-6200
Tel: 301-435-3825
E-mail: hydec@mail.nih.gov
Andrzej Joachimiak, Director
Structural Biology Center & Midwest Center for Structural Genomics
Biosciences Division, Argonne National Laboratory
9700 S. Cass Ave.
Argonne, IL 60439
Tel: 630-252-3926
Fax: 630-252-6991
E-mail: andrzejj@anl.gov
Warren Jones, Ph.D.
Chief
Biochemistry and Bio-related Chemistry Branch
Pharmacology, Physiology, & Biological Chemistry Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2AS.43E
Bethesda, MD 20892-6200
Tel: 301-594-3827
Fax: 301-480-2802
E-mail: jonesw@nigms.nih.gov
Arthur M. Katz, Ph.D.
Physical Scientist
Office of Biological and Environmental Research
U.S. Department of Energy
1000 Independence Avenue, SW
Washington, DC 20585-11290
Tel: 301-903-4932
Fax: 301-903-8521
E-mail: arthur.katz@science.doe.gov
Catherine Lewis, Ph.D.
Director
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.19C
Bethesda, MD 20892-6200
Tel: 301-594-0828
Fax: 301-480-2004
E-mail: lewisc@nigms.nih.gov
J. Jerry Li, M.D., Ph.D.
Program Director
Division of Cancer Biology
National Cancer Institute (NCI)
6130 Executive Blvd, EPN 5004
Rockville, MD 20892-7364
Tel: 301-435-5226
Fax: 301-480-2854
E-mail: jerry.li@nih.gov
Pamela A. Marino, Ph.D.
Program Director
Pharmacology, Physiology, and Biological
Chemistry Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.55E
Bethesda, MD 20892-6200
Tel: 301-594-3827
Fax: 301-480-2802
E-mail: marinop@nigms.nih.gov
Gaetano T. Montelione, Ph.D.
Center for Advanced Biotechnology and Medicine
Structural Bioinformatics Laboratory
Department of Molecular Biology and Biochemistry
Rutgers University and UMDNJ-Robert Wood Johnson
School of Medicine
679 Hoes Lane
Piscataway, NM 08854
Tel: 732-235-5321
E-mail: guy@cabm.rutgers.edu
Arnold Revzin, Ph.D.
Scientific Review Officer
NIH - Center for Scientific Review
Rockledge II, Room 4146
6701 Rockledge Drive
Bethesda, MD 20892-7806 (US Mail)
20817 (courier services)
Tel: 301-435-1153
Fax: 301-480-2327
E-mail: revzina@csr.nih.gov
Michael E. Rogers, Ph.D.
Director
Pharmacology, Physiology, and Biological Chemistry Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.49C
Bethesda, MD 20892-6200
Tel: 301-594-3827
Fax: 301-480-2802
E-mail: rogersm@nigms.nih.gov
Chris Sander, Ph.D.
Computational Biology Center
Memorial Sloan-Kettering Cancer Center
1275 York Avenue, Box 460
New York, NY 10021
Tel: 646-888-2602
Fax: 212-214-0744
E-mail: sanderc@mskcc.org
Dr. Salvatore Sechi
Director, Proteomic Program
Division of Diabetes, Endocrinology, and Metabolic Diseases
National Institute of Diabetes and Digestive and Kidney Diseases
6707 Democracy Blvd. Room 611
Bethesda, MD 20892-5460
Tel: 301-594-8814
Fax: 301-480-3503
E-mail: Salvatore_Sechi@nih.gov
Paul A. Sheehy, Ph.D.
Deputy Associate Director for Extramural Activities
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2AN.24G
Bethesda, MD 20892-6200
Tel: 301-594-4499
Fax: 301-480-1852
E-mail: sheehyp@mail.nih.gov
Ward W. Smith, Ph.D.
Program Director
Cell Biology and Biophysics Division
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.19F
Bethesda, MD 20892-6200
Tel: 301-443-9375
Fax: 301-480-2004
E-mail: smithwar@nigms.nih.gov
Terry R. Stouch, Ph.D.
6211 Kaitlyn Court
West Windsor, NJ 08550
Tel/Fax: 609-275-7234
E-mail: tstouch@gmail.com
Helen R. Sunshine, Ph.D.
Chief, Office of Scientific Review
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 3An.12F
Bethesda, MD 20892-6200
Tel: 301-594-2881
Fax: 301-480-8506
E-mail: sunshinh@nigms.nih.gov
Amy L. Swain, Ph.D.
Biomedical Technology Division
National Center for Research Resources
National Institutes of Health
6701 Democracy Blvd.
Room 964, MSC 4874
Bethesda, MD 20892-4874
Tel: 301-435-0752
Fax: 301-480-3659
E-mail: SwainA@mail.nih.gov
Mona R. Trempe, Ph.D.
Office of Scientific Review
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 3aN.12A
Bethesda, MD 20892-6200
Tel: 301-594-3998
Fax: 301-480-8506
E-mail: trempemo@mail.nih.gov
Jermelina Tupas, Ph.D.
Program Director
Division of Minority Opportunities in Research (MORE)
National Institute of General and Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200
Bethesda, MD 20892-6200
Tel: 301-594-3900
Fax: 301-480-2753
E-mail: tupasjer@nigms.nih.gov
Janna P. Wehrle, Ph.D.
Program Director
Division of Cell Biology and Biophysics
National Institute of General Medical Sciences
National Institutes of Health
45 Center Drive, MSC 6200, Room 2As.19K
Bethesda, MD 20892-6200
Tel: 301-594-5950
Fax: 301-480-2004
E-mail: wehrlej@nigms.nih.gov
MEETING NOTEBOOK
Table of Contents
Future Structural Genomics Initiatives Meeting
October 30-31, 2008
Bethesda, Maryland
- Table of Contents
- Meeting Charge
- Agenda
- Structured Discussion Session Topics
- Roster of Invited Speakers and Other Participants
- Speaker Abstracts and Biosketches
- PSI Investigator's White Paper
- Basic PSI Administrative Information - including Administrative Structures
- Other Background Documents
- Meeting Administrative Materials - Hotel, Dinner, Reimbursement, etc.
Tab 9 - Detail Other Background Documents
- RFAs - Titles and Links
- PSI Assessment Report - Message from Director, NIGMS
- PSI Assessment Report - Janet Smith, Chair
- Commentary on the Assessment Report - Robert Service - Science
- PSI Annual Meeting Summary and Update - Wayne Hendrickson
- PSI Advisory Committee Meeting and Update - Brian Matthews
- PSI Knowledgebase Steering Committee Report - Eaton Lattman
- PSI Knowledgebase Summary - Helen Berman
- PSI Materials Repository Summary - Joshua Labaer
- PSI Knowledge Index - Nature Gateway - Website
- PSI Target Selection Document - Website
- PSI Metagenomic Project - Website
- PSI Framework for Community Nominated Targets - Website
- PSI Workshop on the Biological Annotation of Novel Proteins - Website
- PSI Bottlenecks Workshop - Website
- Protein Production and Purification - Nature Methods - PSI Investigators
- Novel Leverage of Structural Genomics - Nature Biotechnology - Burkhard Rost
- Report of Workshop on Applications of Protein Models in Biomedical Research - Torsten Schwede and Andrej Sali
- Unique Opportunities for NMR Methods in Structural Genomics - Gaetano Montelione, et al.
- The Nature of the Protein Universe - Michael Levitt
- Update on the Protein Structure Initiative - Structure - Norvell and Berg
- Links to other Commentaries in Structure, other editorial pieces, and other relevant papers
- Other Useful Links
- PSI Calendar of Meetings and Events
