Dr. Data
PDF print versionYou will need Adobe Acrobat Reader to view PDFs on this page.
WATCH/LISTEN
"Get excited about biology and computer science!"
—Atul Butte
March 2008 issue
 Video
[MOV, 12.5MB] (Captions)
Video
[MOV, 12.5MB] (Captions)
[Download
free Quicktime Player]
![]() Audio
[MP3, 6.55 MB] (Transcript)
Audio
[MP3, 6.55 MB] (Transcript)
GET MORE INFO
National
Library of Medicine’s Online Fact Sheet on Microarrays http://www.ncbi.nlm.nih.gov/About/primer/microarrays.html.
Animation:
DNA Microarray Methodology
Tutorial:
NHGRI - Bioinformatics
Tutorial:
NHGRI - How to Sequence a Human Genome
Atul Butte is one of those people who thinks big. Really big.
Trained as both a doctor and a researcher, he's always busy with something—he does gene experiments on his computer one day and sees patients another. But Butte isn’t satisfied with studying just one disease or a single gene.
Instead, he throws his net over all of them. Then he reels in tantalizing patterns that take shape in the vast sea of data.
Butte, 38 (whose name is pronounced Byoot), is both a pediatrician and a bioinformatics researcher at Stanford University in California. He is using computers to analyze the activity of all 20,000-plus genes in the human genome.
Specifically, Butte is examining which genes rev up and which stall in diabetes, heart disease, muscular dystrophy, or other conditions. That, he hopes, will lead to a new understanding of diseases and new ways of treating them.
"The overall goal of my work is to redefine our entire knowledge about all diseases and to predict new uses for all existing drugs," he says.
That may seem like an incredibly ambitious dream, but if you met Butte, you’d understand. His energy, zeal—and lots of coffee—keep him in nearly constant motion.
Career Crisis
Butte has always had twin passions—medicine and computers. Although he didn't realize it until he was nearly done with his schooling, feeding both of his passions was the perfect preparation for his eventual career.
He has wanted to be a doctor as long as he can remember. After high school, Butte enrolled in a program at Brown University in Providence, Rhode Island, that allows college students to major in virtually anything—and guarantees that they get into Brown’s medical school.

Dr. Atul Butte
Courtesy: Steve Gladfelter
But rather than most pre-med students, who pick majors like biochemistry in college, Butte chose computer science. He got summer jobs in computer programming, working at Apple, Microsoft, and other computer firms.
Meanwhile, Butte had 4 years of medical school ahead of him, and then even more years after that if he wanted to become a specialist.
“I had a bit of crisis,” Butte admits. “I didn’t know whether to just bail and go into the computer industry.”
Ultimately, medicine won. Butte decided he’d rather care for patients and advance the field of medicine than build tools to let other people do it.
Focus Found
While Butte was in medical school, he saw the chance to ride another wave of excitement. Now, it was the life sciences that were taking off, thanks to the Human Genome Project and other technological wonders just coming online.
He was lucky to find an opportunity to see first-hand what all of the hubbub was about. He got a spot in a year-long program sponsored by the Howard Hughes Medical Institute. The program allows medical students to work side by side with laboratory researchers at the National Institutes of Health in Bethesda, Maryland.

Butte created this tapestry image using 8.4 million measurements
of human gene activity (light/dark lines).
Courtesy: Atul Butte, NASA (Earth)
What Butte learned that year steered the rest of his career. He discovered that, although biological questions fascinated him, he wasn’t suited for bench research (“I was terrible in the lab,” he claims).
Even more significant, he was introduced to a topic that completely captivated him and cemented his interest in one disease in particular: diabetes.
“I hadn’t even thought about diabetes before,” Butte says, although “this disease affects more than 10 percent of the world’s population.”
Butte explains that while thousands of researchers are working hard on the problem, much remains a mystery.
“We still don’t even know why people get diabetes,” Butte says.
Butte decided to go into pediatric endocrinology. Doctors in this specialty deal with the body’s production and use of insulin and other hormones. Butte treats children with diabetes and growth problems.
He says he chose pediatrics “because it’s the field of medicine that’s closest to genetics.” That’s because many serious childhood diseases can result from genetic, rather than environmental, causes.
All the pieces came together when Butte met Isaac (“Zak”) Kohane, a scientist at Harvard Medical School in Boston, Massachusetts, and began working with him. Kohane was a perfect mentor for Butte: Like Butte, Kohane is a pediatric endocrinologist with an interest (and a Ph.D.) in computer science.
Spending time in Kohane’s lab, Butte learned to balance medicine and computation in an integrated and synergistic way. He also earned master’s and doctoral degrees in medical informatics, a field that blends information science with the analysis of medical data.
The experience landed Butte a dream job at Stanford, where he now combines his two passions in a cutting-edge career.
Dusting Off Disease Definitions

Inspired by old books like this one, Butte wants to modernize the way we think about diseases.
According to Butte, the way we think about diseases is antiquated. And he’s become a bit of an expert on the subject.
Thanks to Google™ Book Search, Butte developed an interest in history—especially in the history of disease classification, or nosology. One of the books he uncovered provides code numbers for each of the commonly recognized causes of death in 1909.
Cancers are listed as code numbers 39 to 45 and include cancer of the oral cavity, stomach, liver, peritoneum, intestines, rectum, female genital organs, breast, skin, and “other organs.”
“Imagine!” he exclaims. “Lung cancer, which now kills more Americans than any other cancer, was lumped in with ‘other organs.’ And that was less than 100 years ago!”
Butte thinks that the way we classify diseases is horribly obsolete and that the way we treat them is too.What he wants to do is modernize nosology by replacing our current, anatomy—and symptoms—based system with one based on information from genes.
According to Kohane, if anyone can pull this off, Butte can.
“He’s absolutely meticulous and exhaustive.”
A Genomic Surprise

Surprise! Gene studies suggest that the hyrax is the closest living
relative to an elephant. Butte expects to uncover genomic surprises
about health and disease.
Courtesy: Dave Mangham
Can old medicines learn new tricks? Butte thinks so.
His ultimate goal in creating a genome-based disease classification system is to come up with new uses for existing medicines. And Butte may already have almost succeeded by finding a surprising connection between heart attacks and muscular dystrophy.
The two diseases could hardly look more different, Butte says.
Heart attacks typically affect older people after decades of accumulated damage to blood vessels. Muscular dystrophy, on the other hand, appears in the toddler years as progressive muscle weakness. The disease is incurable, and patients die in their teens or early 20s.
Yet, according to Butte, heart attacks and muscular dystrophy are pretty similar at the genomic level. In other words, both diseases alter the activity of the same group of genes.
So, could the same medicines treat both diseases? Butte hopes so.
Currently, there are more than 40 medicines used to treat heart attacks but only one for muscular dystrophy, and it’s not a cure.
If existing heart-attack drugs turn out to work against muscular dystrophy, not only would this provide immediate benefits for the thousands of children with the disease, it would also be a huge savings in time and money.
That’s because a pharmaceutical company typically spends close to $1 billion and more than 10 years to develop a new medicine from scratch.
The Power of GEO
Much of the data that Butte taps into is in the form of microarrays, which are also called gene chips or DNA chips. That’s because they are often manufactured like computer chips.
The thumbnail-sized devices are microscopic grids that have pieces of DNA representing every gene in the human genome stuck on them. Scientists use them to measure the activity of our 20,000-plus genes at the same time.
By using multiple microarrays, scientists can compare how patterns of gene activity change under different conditions, like in different diseases. Butte compared the patterns of gene activity in people with muscular dystrophy to those of people who survived a heart attack.
This approach—analyzing mountains of data simultaneously to find meaningful patterns and tantalizing surprises—is a radical departure from the way most medical research has been done for the past 20 or so years, with an intense focus on one gene, one protein, or one biological process.
Researchers like Butte who want to publish scientific studies using microarrays are required to deposit their data into databases like GEO, the Gene Expression Omnibus. The data is coming hard and fast: Scientists are submitting more than 1,000 files each week.
That means that by the time this article is published, GEO will contain the digitized microarray results from more than 200,000 samples.

Scientists use DNA chips like this one to measure gene activity.
All the data in GEO is freely available online, enabling anyone with a reasonably good personal computer and an Internet connection to do bioinformatics experiments (see “How You Can Be a Bioinformatician”).
“So any high school student who wants to do an experiment for a science-fair project can start with some 200,000 microarrays—and that number is doubling or tripling each year,” says Butte.
“Isn’t this an amazing world?”
But there’s a downside to this wealth of information: Butte and other scientists are drowning in data.
“Life-science data is growing faster than computational power,” he says. “It’s outpacing what hard drives and processors can do.”
As a result, Butte can’t simply use brute force to tackle the research topics that interest him—he has to choose his questions very wisely.
To do that, Butte focuses on “big” questions about health—questions that can only be answered by analyzing hundreds of diseases and tens of thousands of genes simultaneously.
Like these:
What are the molecular similarities and differences across all diseases that plague us?
Which genes are affected in every single human disease?
Can we find biological markers that predict diseases before symptoms show up?
Can we use the molecular similarity between diseases to help us apply drugs for one disease to another?
“Those are the kind of cool questions that no one could answer before,” Butte says.
The Good Things in Life
Although intensely committed to science and medicine, Butte’s job is not the only driving force in his life.
“Before he got married,” Kohane says, “he used to work all the time at the hospital—all hours of the day and night.”
Now, Butte tries to get home in time for dinner and loves to talk about the latest amusing or impressive things that his daughter is doing.
“He’s a family man, and I really respect that,” Kohane says.
Butte is also well-known for being a coffee connoisseur.

Butte fuels his enthusiasm with lattes and other coffee drinks.
Because of their WiFi connections (and caffeine), Butte was an early fan of Starbucks® coffee shops. He even had his first date with his wife there.
Butte is one of those people who orders incredibly complex coffee drinks.“He has completely mastered the idiosyncratic language Starbucks uses to describe their coffees,” Kohane laughs.
Butte’s favorite is an iced grande, non-fat, no-whip, raspberry mocha.
Butte also loves good food. “Whenever I’d meet him in any part of the world,” Kohane says, “he’d always know where all the best restaurants were in detail, including their closing times and ratings.”
Could it be that Butte is getting a bit of help from technology? Indeed.
During a recent meeting in Vienna, Austria, for example, Butte and a coworker had a digital duel to find directions to a restaurant.
Each whipped out his electronic sharpshooter—Butte has an HTC TyTN smartphone with a 3G UMTS/HSDPA wireless connection. His friend had an iPhone with a 2.5 G EDGE wireless connection.
Butte won. By a lot.
Of course, as a bioinformatician, Butte also uses gadgets professionally.
In his research lab, he uses a networked cluster of 64 hyper-threaded CPUs to crank away at staggeringly complicated computational problems.
And for nearly a decade, he has downloaded and indexed on his laptop every research article relevant to his scientific interests.
“When attending seminars, he’ll ask questions quoting these papers as if out of an infinite knowledge base,” says Kohane.
Passion Level High, Prognosis Good
For Butte, though, gadgets will never be enough by themselves. He wants his computational findings to reach the people who need them—real kids and real adults with real diseases.
“Life-science data is growing faster than computational power.”
“When I first met [Atul], he was very impatient with the current state of medicine, and that really made him stand out,” remembers Kohane. “It was clear that he wanted to help cure the diseases that affect lots of people and that he was going to use all his energies to make it happen.”
Butte’s enthusiasm is infectious, says Kohane. “He draws many people towards him. He’s always smiling and genuinely excited. He’s energized by the potential opportunity to improve the system.”
As things look now, the prognosis is good that Butte will improve many lives, as he develops a recipe for health that perfectly combines computers and compassion.
How You Can Be a Bioinformatician
Try your own bioinformatics experiment! Explore links between cigarette smoking and lung cancer, go to http://www.ncbi.nlm.nih.gov/geo/.
Type “cigarette smoke” into the second empty box, which is next to the “Gene profiles” label, and click GO. You’ll get information on thousands of genes. Look for the experiment (numbered row) that says “UBE2D3: ubiquitin-conjugating enzyme E2D 3.”
In this experiment, the researchers studied human lung cells grown in the lab. Some of the cells were exposed to 15 minutes of cigarette smoke, and others (the controls) were kept in a smoke-free environment.
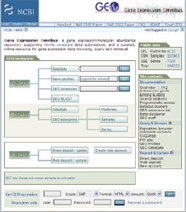 Courtesy: GEO/NCBI/NIH
Courtesy: GEO/NCBI/NIH Click on the chart to the right to see an enlarged version. The left half of the chart shows the activity of the UBE2D3 gene in control cells. The right half shows the gene’s activity in cells exposed to smoke. The height of the red bars shows how active this gene is. As you can see, UBE2D3 activity increases noticeably in response to cigarette smoke.
To get a list of other genes that also activate in response to cigarette smoke, go back one page and click on “Profile Neighbors” above the chart. Then scroll down to see the charts of UBE2D3’s “neighbors.” You’ll see that the patterns in all the charts look much the same, with mostly red bars on the right side of the graph.
Because these genes ramp up when exposed to cigarette smoke, some may be linked to lung cancer. Genes that are activated whether or not smoke is present are probably unrelated to lung cancer, and these do not show up as “neighbors” to the UBE2D3 gene.
In this simple search, you uncovered several genes that could help researchers learn more about how lung cancer develops and how to design drugs against the disease.
For an example of bioinformatics in action, go to the presentation called “Aging and the Human Brain.”at http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=coffeebrk.box.666 —A.Z.M.

