| |

“The findings and conclusions in this review are those of the author(s)
and do not necessarily
represent the views of the funding agency.” |
This HuGE Review was published in Am J of Epidem 2006; 164(8):711-724 |
|
Sun-Wei Guo, Victoria L. Magnuson, Jennifer J. Schiller, Xujing Wang, Yan Wu and Soumitra Ghosh
From the Department of Pediatrics and the Max McGee Research Center for Juvenile Diabetes, Medical College of Wisconsin, Children's Research Institute, Children's Hospital of Wisconsin, Milwaukee, WI Correspondence to Dr. Sun-Wei Guo, Department of Pediatrics, Medical College of Wisconsin, 8701 Watertown Plank Road, MS 756, Milwaukee, WI 53226-0509 (e-mail: swguo@mcw.edu). Received for publication January 17, 2006. Accepted for publication March 31, 2006.
|
ABSTRACT |
Several polymorphisms in the vitamin D receptor (VDR) gene have been reported to be associated with the risk of developing type 1 diabetes, yet published findings have been conflicting. In this study, the authors attempted to evaluate the evidence regarding the association. They searched all relevant reports from original papers published from 1997 to December 2005. Predefined criteria were used to identify 1) case-control association studies examining the FokI (11 studies), BsmI (13 studies), ApaI (9 studies), and TaqI (7 studies) polymorphisms and 2) a few family-transmission studies with analysis of these four polymorphisms. In random-effects modeling, the 95% confidence intervals of the summary odds ratios for all four polymorphisms included 1, indicating no effect. Except for FokI, no heterogeneity was found. The 95% confidence intervals of the transmission proportions all included 0.5, indicating no effect. Thus, the authors found no evidence for an association between VDR gene polymorphisms and type 1 diabetes risk in either case-control studies or family-transmission studies. In fact, a reanalysis of previously published data (McDermott et al., Diabetologia 1997;40:971–5) indicated no evidence of an association as reported.
Keywords: association; diabetes mellitus, type 1; epidemiology; genetics; meta-analysis; polymorphism, genetic; receptors, calcitriol; VDR

|
INTRODUCTION |
That type 1 diabetes has a strong genetic component is now indisputable. Human leukocyte antigen class II genes have been identified as the most important genetic factor in determining the risk of developing type 1 diabetes. The VNTR (variable number of tandem repeats) polymorphism located in the promoter region of the insulin gene and the cytotoxic T-cell-associated antigen-4 gene also have been identified (1, 2). However, these genes are neither sufficient nor necessary to cause type 1 diabetes. Hence, the search for other genes and environmental triggers has been ongoing.
Investigators from several epidemiologic studies have reported that dietary vitamin D supplementation during infancy and childhood reduces the risk of type 1 diabetes (3–5). Recently, in the Diabetes Autoimmunity Study in the Young, Fronczak et al. (6) reported that the presence of islet autoantibodies in offspring was inversely correlated with maternal dietary vitamin D intake during pregnancy. Vitamin D receptor (VDR) polymorphisms are reported to be associated with insulin secretory capacity in humans (7). In experimental studies, oral administration of 1 ,25-dihydroxyvitamin D3, the activated form of vitamin D, completely protects NOD mice from type 1 diabetes (8, 9). In addition, it has been reported that a vitamin D analog can down-regulate proinflammatory chemokine production by pancreatic islets, thereby inhibiting T-cell recruitment and development of type 1 diabetes (10). These epidemiologic and experimental data appear to indicate that vitamin D deficiency may be involved in the pathogenesis of type 1 diabetes, possibly because vitamin D is a potent modulator of the immune system and is involved in regulating cell proliferation and differentiation (11, 12). ,25-dihydroxyvitamin D3, the activated form of vitamin D, completely protects NOD mice from type 1 diabetes (8, 9). In addition, it has been reported that a vitamin D analog can down-regulate proinflammatory chemokine production by pancreatic islets, thereby inhibiting T-cell recruitment and development of type 1 diabetes (10). These epidemiologic and experimental data appear to indicate that vitamin D deficiency may be involved in the pathogenesis of type 1 diabetes, possibly because vitamin D is a potent modulator of the immune system and is involved in regulating cell proliferation and differentiation (11, 12).
Vitamin D and its analogs exert their actions through VDR, which is a member of the steroid hormone receptor superfamily. The VDR gene, located on chromosome 12q12–q14, has at least five promoter regions (13), eight protein-coding exons, and six untranslated exons, which are alternatively spliced (14). FokI (in exon 2), BsmI and ApaI (both in intron 8), and TaqI (in exon 9) are the four common single nucleotide polymorphisms (SNPs) (rs10735810, rs1544410, rs7975232, and rs731236
, respectively) in the VDR gene that have been most often investigated (12, 15).
The first report of a type 1 diabetes-VDR association was made by McDermott et al. (16) in 1997. They reported that the "b" allele of the BsmI polymorphism in the VDR gene was preferentially transmitted to offspring afflicted with type 1 diabetes. As with other genetic association studies, however, reports on the type 1 diabetes-VDR association have been conflicting. A recent study involving over 3,000 families with type 1 diabetes found no evidence for an association of type 1 diabetes with any of the four SNPs mentioned above or with any of numerous other polymorphisms across the VDR gene (17).
Since an individual study may not have enough statistical power to detect any association between type 1 diabetes and VDR polymorphisms, a meta-analysis that combines data from all published studies may provide a more accurate estimate of effect sizes, leading to a reduced probability of false-negative results (18). Thus, we conducted a comprehensive and quantitative assessment of the association between type 1 diabetes and the four aforementioned polymorphisms. We sought to estimate effect sizes and to determine the extent of heterogeneity in the strength of associations between studies.

|
MATERIALS AND METHODS |
| Search strategy and inclusion/exclusion criteria
We searched the US National Library of Medicine's PubMed database in a systematic and diligent manner for all genetic association studies on VDR and type 1 diabetes published from 1997, when the VDR-type 1 diabetes association was first reported (16), through December 2005. We focused on the four most-studied polymorphisms: ApaI, BsmI, FokI, and TaqI. The search used the keywords "association studies," "insulin-dependent diabetes," "type 1 diabetes," "vitamin D receptor," "ApaI," "BsmI," "FokI," "TaqI," and "polymorphisms," as well as combinations thereof. The references of all computer-identified publications were searched for additional studies, and the PubMed option "Related Articles" was used to search for potentially relevant papers. Papers published in any language were selected if they met the following criteria: 1) the publication was an association study, either of the case-control type or a family-transmission study, and 2) the publication reported genotypic frequencies of VDR polymorphisms in unrelated type 1 diabetes patients and unrelated individual controls, or reported proportions of transmission of specific alleles at a locus in the VDR gene. In the case of sequential or multiple publications of analyses of the same data or overlapping data sets, the publication that reported data from the largest or most recent study was included, as recommended by Little et al. (19).
Data extraction
Following the MOOSE (Meta-analysis Of Observational Studies in Epidemiology) guidelines for reporting on meta-analyses of observational studies (20), the following data were extracted from the eligible studies: authors' names; region/country where the study was conducted; year of publication; numbers of cases/patients and controls or number of families studied; mean age (or range) at onset of type 1 diabetes in cases/patients or probands; diagnostic criteria; mean age (and standard deviation) or age range in the control group; manner in which the controls were selected; and number of subjects with the VDR genotype in both cases and controls. Information on whether the investigators had made any attempt to test for Hardy-Weinberg proportion in the controls, to check for and correct genotyping errors, and to control for confounding risk factors was also noted.
The search produced 21 published papers on the genetic association between the VDR gene and type 1 diabetes, 20 in English (7, 16, 17, 21–38) and one in Spanish (39). Data used in the article by Marti et al. (39) appeared to be identical to the data reported by Audi et al. (32), so the paper by Marti et al. was excluded. Eerligh et al. (31) did not report the transmission probability of any VDR polymorphism, and thus this paper was also excluded. The more detailed genotypic data not presented in the paper by Yokota et al. (28) were kindly provided by Dr. Yokota and were included. Turpeinen et al. (35) reported data on populations from three different geographic regions; these data were counted as three different studies in this analysis. For similar reasons, data from two different areas (Barcelona and Navarre) in the paper by Audi et al. (32) were counted as two separate studies. Therefore, of the 21 papers identified, two were excluded. Of the 19 remaining papers, six were family studies reporting numbers of transmitted alleles or haplotypes at ApaI, BsmI, FokI, and/or TaqI in type 1 diabetes patients (16, 17, 24, 25, 34, 37), and the rest reported genotype/haplotype frequencies at FokI, BsmI, ApaI, and/or TaqI in cases and controls. In all cases, genotypes were designated by a lowercase letter for the presence of a restriction site and a capital letter for its absence.
Statistical analysis
The goals of this analysis were to pool crude odds ratio estimates from included case-control studies, to identify any heterogeneity, and, if heterogeneity was present, to attempt to identify its sources. For each included study, we computed the odds ratio for having type 1 diabetes in people with the restriction site (lowercase letter) as compared with people without the site (uppercase letter) and its 95 percent confidence interval, as well as the standard error of the log odds ratio. We chose the log odds ratio simply because, unlike the odds ratio, its standard error is unaffected by its magnitude. For family studies, we calculated a summary of transmission proportions, weighted by the inverse of the estimated variance of the transmission proportion of each study.
Since all four of the studied SNPs (FokI, BsmI, ApaI, and TaqI) are diallelic, we calculated summary odds ratios incorporating both within- and between-study variation, using a random-effects model proposed by DerSimonian and Laird (40). This model provides a means of testing for heterogeneity in odds ratio estimates across studies. The random-effects model stipulates that, in addition to sampling errors, the studies may have genuine variations in their results attributable to some unknown variables not accounted for by all studies.
For gene transmission data, we calculated the pooled estimate of the transmission proportion by using the inverse of the variance of the individual estimates as weights. The test for heterogeneity was carried out using the method of Potthoff and Whittinghill (41).
Funnel plots were used to examine asymmetry, in which the odds ratios were plotted on a logarithmic scale against the inverse of their corresponding standard errors, a measure of precision (42). The funnel plots were also used to plot the transmission proportions from each study against the inverse of their standard errors. If bias is absent, small studies will have odds ratios or transmission proportions that are widely scattered but symmetric about the odds ratio or the transmission proportion estimates provided by larger, more precise studies. In this case, the plot would resemble a funnel with the tip pointing approximately towards the true log odds ratio or the transmission proportion. If publication bias is present, the plot will be asymmetric, since some studies with negative findings are not published.
To examine whether the association between a particular allele at a given locus and type 1 diabetes was genuine rather than an artifact of multiple comparisons, we used funnel plots to search for frequency differences in specific alleles between cases and controls. Specifically, we plotted the difference in allele/genotype frequency, pi – qi, versus its standard error. When the odds ratio was of interest, the log odds ratio versus its standard error was plotted. When no allele is associated with type 1 diabetes, the plot should resemble a funnel, with the tip of the funnel pointing to 0. When a particular allele/genotype is associated with type 1 diabetes, the frequency difference of that particular allele/genotype should conspicuously stand out. To control for multiple comparisons when testing for differences in k genotypes of a gene between cases and controls, we used the resampling procedure described by Westfall and Young (43) for computing adjusted p values.
We also employed funnel plots to examine whether a particular haplotype was genuinely preferentially transmitted to offspring with type 1 diabetes rather than an artifact of multiple comparisons, where the precision was defined as the standard error of the transmission proportion. The p value for excessive transmission was calculated under the null hypothesis that the transmission proportion was 50 percent. Since different haplotypes are transmitted independently, we used the method of Benjamini and Hochberg (44) to control for multiple comparisons with a 5 percent false-discovery rate.
All computations were carried out in R, version 2.2.0 (www.r-project.org). 

|
RESULTS |
Characteristics of included studies
In the 19 published papers included in the meta-analysis, the BsmI, ApaI, and TaqI polymorphisms had been investigated in both case-control studies and family studies. Nine studies on the ApaI-type 1 diabetes association recruited 1,101 cases/patients and 2,805 controls, while 13 studies on the BsmI polymorphism recruited 1,601 cases/patients and 4,207 controls. For the FokI polymorphism, 11 studies included 1,424 cases and 3,301 controls, while seven studies on the TaqI polymorphism included 681 cases and 781 controls. Detailed characteristics of each study, along with p values for testing Hardy-Weinberg proportion, are listed in Tables 1–4 for FokI, BsmI, ApaI, and TaqI, respectively. The characteristics of all of the family studies are listed in Table 5.
|
|
Table 1: Characteristics of case-control studies included in a meta-analysis of the relation between the FokI polymorphism in the vitamin D receptor gene and type 1 diabetes |
|
|
Table 2: Characteristics of case-control studies included in a meta-analysis of the relation between the BsmI polymorphism in the vitamin D receptor gene and type 1 diabetes |
|
|
Table 3: Characteristics of case-control studies included in a meta-analysis of the ApaI polymorphism in the vitamin D receptor gene and type 1 diabetes |
|
|
Table 4: Characteristics of case-control studies included in a meta-analysis of the TaqI polymorphism in the vitamin D receptor gene and type 1 diabetes |
|
|
Table 5: Characteristics of family studies included in a meta-analysis of various polymorphisms in the vitamin D receptor gene and type 1 diabetes |
The frequency of the "f " allele at FokI among controls ranged from 56.3 percent in Hungarians to 68.8 percent in the Basque population of Spain. The frequency of the "a" allele at ApaI among controls ranged from 26.4 percent in Chinese in Taiwan to 63.6 percent in Croatians, while the frequency of the "b" allele ranged from 3.6 percent in Taiwan Chinese to 53.5 percent in Germans. The frequency of the "t" allele at TaqI ranged from 50.9 percent (Germans) to 96.8 percent (Taiwan Chinese).
For family studies, the number of families studied ranged from 59 in the paper by Angel et al. (34) to over 3,000 in the paper by Nejentsev et al. (17).
Diagnosis of type 1 diabetes
Type 1 diabetes is often diagnosed mainly on the basis of clinical symptoms alone, which include malaise, weight loss, thirst, and polyuria. Insulin is required soon after diagnosis, and ketosis is common. The presence of autoantibodies (to glutamic acid decarboxylase 65, islet antigen 2, and insulin) adds to confidence in the diagnosis, while elevated C-peptide levels are not normally required for diagnosis. Of the articles surveyed here, only eight (22–24, 26, 29, 34–36) used the World Health Organization criteria for type 1 diabetes, which in essence are captured by our description above. Investigators in five papers (16, 23, 27, 30, 35) specifically looked for the presence of autoantibodies to distinguish type 1 diabetes from type 2 diabetes. In the rest of the papers, investigators used only one criterion (e.g., ketosis, early requirement for insulin) to diagnose type 1 diabetes.
Qualitative assessment of included studies
Most studies focused on juvenile type 1 diabetes, as seen by the age ranges of the cases/probands selected (Tables 1–5). For case-control-type association studies, the selection of cases or patients varied: Some investigators selected prevalent cases, while others selected incident cases. In the earlier studies, researchers tended to pay less attention to methodological details, such as the criteria used for case/patient selection and age.
The selection of controls varied substantially. Some groups of controls were population-based while others were hospital-based, and both appeared to be selected on the basis of convenience. For population-based controls, for example, terms such as "donors" or "racially matched controls" were used. For hospital-based studies, controls ranged from healthy children undergoing check-ups to healthy infants without type 1 diabetes enrolled in an intervention trial.
Despite such heterogeneity, there were several features shared by all case-control studies. First, no information was provided in any surveyed study regarding the assessment of Hardy-Weinberg proportion in controls. Since assessment of Hardy-Weinberg proportion is regarded as an important criterion in the evaluation of genetic association studies (19), caution should be exercised in interpreting these studies. Second, in no study was an attempt made to match controls with the cases on factors that may have confounded the results. For example, human leukocyte antigen is by far the only genetic factor consistently reported to be associated with type 1 diabetes, yet no study attempted to match control human leukocyte antigen haplotypes with those of cases. In some studies, the controls as a group were apparently younger than the cases, leaving open the possibility that some of them may have developed type 1 diabetes later in life. Lastly, none of the studies made much of an attempt to control for known risk factors for type 1 diabetes, such as dietary intake of vitamin D during infancy and/or childhood.
The FokI, BsmI, ApaI, and TaqI polymorphisms and the risk of type 1 diabetes
Case-control studies.
The odds ratio for type 1 diabetes for the "f " allele at FokI ranged from 0.71 to 1.58 (Figure 1, upper left panel). The random-effects model yielded a pooled odds ratio of 1.05 (95 percent confidence interval (CI): 0.89, 1.25). There was some indication of mild heterogeneity (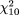 = 18.65, p = 0.045; estimated variance of random effect: = 18.65, p = 0.045; estimated variance of random effect:
 2 = 0.04). 2 = 0.04).
For BsmI, the odds ratio ranged from 0.55 to 13.49 (Figure 1, upper right panel). The random-effects model yielded a pooled odds ratio of 1.09 (95 percent CI: 0.88, 1.36). Again, there was no sign of heterogeneity (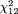 = 15.13, p = 0.23, = 15.13, p = 0.23,  2 = 0.03). For ApaI, the odds ratio ranged from 0.62 to 2.27 (Figure 1, lower left panel). The pooled summary odds ratio based on a random-effects model was 1.11 (95 percent CI: 0.90, 1.37). There was no indication of heterogeneity among studies ( 2 = 0.03). For ApaI, the odds ratio ranged from 0.62 to 2.27 (Figure 1, lower left panel). The pooled summary odds ratio based on a random-effects model was 1.11 (95 percent CI: 0.90, 1.37). There was no indication of heterogeneity among studies (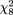 = 12.83, p = 0.12, = 12.83, p = 0.12,  2 = 0.04). 2 = 0.04).
For TaqI, the odds ratio ranged from 0.93 to 1.47 (Figure 1, lower right panel). The random-effects model yielded a pooled odds ratio of 1.02 (95 percent CI: 0.86, 1.20). There was no sign of heterogeneity (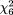 = 2.08, p = 0.91, = 2.08, p = 0.91,  2 = 0.00). 2 = 0.00).
In view of these summary estimates, there is no evidence that any of the four SNPs alone is associated with type 1 diabetes.
Family transmission studies.
For the "f" allele at FokI, the pooled estimate of the transmission proportion was 0.490 (95 percent CI: 0.474, 0.507), with a strong indication of heterogeneity (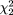 = 9.64, p = 0.008). = 9.64, p = 0.008).
The pooled estimate of the transmission proportion of the "a" allele at the ApaI locus from six studies (Table 5) was 0.498 (95 percent CI: 0.484, 0.512). There was no indication of heterogeneity (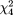 = 4.68, p = 0.32). For the "t" allele at TaqI, the pooled estimate of the transmission proportion was 0.496 (95 percent CI: 0.479, 0.514). Heterogeneity did not appear to be present ( = 4.68, p = 0.32). For the "t" allele at TaqI, the pooled estimate of the transmission proportion was 0.496 (95 percent CI: 0.479, 0.514). Heterogeneity did not appear to be present (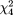 = 2.44, p = 0.65). For the BsmI polymorphism, we used the numbers reported by McDermott et al. (16); that is, the "b" allele was considered to have been transmitted 53 out of 84 times, although Table 2 in the published article (16) gave the number as 75/132. The pooled estimate of the transmission proportion was 0.490 (95 percent CI: 0.473, 0.506). There was no indication of heterogeneity ( = 2.44, p = 0.65). For the BsmI polymorphism, we used the numbers reported by McDermott et al. (16); that is, the "b" allele was considered to have been transmitted 53 out of 84 times, although Table 2 in the published article (16) gave the number as 75/132. The pooled estimate of the transmission proportion was 0.490 (95 percent CI: 0.473, 0.506). There was no indication of heterogeneity (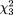 = 7.26, p = 0.06). = 7.26, p = 0.06).
The funnel plot of the transmission proportions of the "b" allele from four studies is depicted in Figure 2. The plot shows that as the sample size increases, the transmission proportion gravitates towards the equal transmission proportion of 0.5. Funnel plots for FokI, ApaI, and TaqI produced similar patterns (data not shown). Therefore, there was little indication that any of the four polymorphisms showed preferential transmission to affected offspring.
Combined haplotypes and the risk of type 1 diabetes
For transmission of haplotypes, data were available from McDermott et al. (16), Pani et al. (24), and San-Pedro et al. (38) (Tables 6 and 7). McDermott et al. reported that the "b" allele was preferentially transmitted to affected offspring (53 of 84 times; p = 0.016) and that the bAT haplotype was transmitted as well (35 of 54 times; p = 0.0295) (16). Interestingly, an entirely different transmission proportion for the "b" allele could be deduced from McDermott et al.'s Table 2 (reproduced in Table 7 here): 0.568 (75/132 = 0.568; p = 0.069 in an exact test based on the binomial distribution), which was not significant at the 5 percent level. Using the procedure of Benjamini and Hochberg (44) with a 5 percent false-discovery rate, we did not find any evidence that the haplotype bAT was preferentially transmitted to affected offspring (Table 6). The funnel plot appeared to indicate that the extent to which the transmission proportion for the bAT haplotype deviated from the null was no more pronounced than that for BAT (Figure 3, left panel).
|
|
Table 6: Numbers of vitamin D receptor gene BsmI-ApaI-TaqI haplotypes transmitted and not transmitted to offspring with type 1 diabetes as reported in studies by McDermott et al. (16) and Pani et al. (24), along with unadjusted p values for transmission and their significance after adjustment for multiple comparisons (false-discovery rate = 0.05)* |
|
|
Table 7: Numbers of vitamin D receptor gene FokI-BsmI-ApaI-TaqI haplotypes transmitted and not transmitted to offspring with type 1 diabetes as reported in a study by San-Pedro et al. (38), along with unadjusted p values for transmission and their significance after adjustment for multiple comparisons (false-discovery rate = 0.05)* |
Pani et al. (24) reported that the haplotype BAt conferred the highest risk of type 1 diabetes. The funnel plot did not appear to dispute this claim (Figure 3, right panel). However, with a family-wise false-discovery rate of 5 percent, the evidence for preferential transmission of the BAt haplotype disappeared (Table 6).
San-Pedro et al. (38) did not find a transmission distortion of the three-locus haplotype examined by McDermott et al. (16) and Pani et al. (24) (Table 2 in the published article (38)). They did report, however, that one four-locus haplotype, fBAt, was transmitted more frequently to offspring with type 1 diabetes (Table 7). Although this distortion was still significant after adjustment for multiple testing, the result appeared to be incongruent with other studies. The funnel plot indicated that it did not deviate much from the funnel shape (Figure 4).
Revisiting the evidence presented in the first report
Some inconsistencies in the first report on the VDR-type 1 diabetes association (16), as mentioned above, prompted us to reexamine the data presented by McDermott et al. According to Table 1 in the paper by McDermott et al. (16), the proportions of transmitted BsmI-TaqI haplotypes were as follows: BT, 11/38; bT, 41/59; Bt, 21/50; and bt, 2/3. This led to the conclusion that the bT haplotype was preferentially transmitted to affected offspring (16). However, the three-locus haplotype data in McDermott et al.'s Table 2 (16) were more informative than the two-locus data, and the proportions of transmitted BsmI-TaqI haplotypes could be deduced to be the following: BT, 15/46; bT, 69/116; Bt, 23/48; and bt, 6/16 (these are the minimum numbers, since in some cases the three-locus haplotype or its transmission cannot be determined but that of the two-locus haplotype can). Transmissions increased 51 percent, from 150 for the BsmI-TaqI system to 226 for the BsmI-ApaI-TaqI system, with the same family data. The two-locus haplotype data deduced from the three-locus data yielded a transmission proportion for the bT haplotype of 0.595 (69/116 = 0.595), which had a p value of 0.025 using a binomial distribution under the null hypothesis. Since four tests have been performed for the four haplotypes, the result was not significant with a 5 percent false-discovery rate. Taken together, it appears that claims made in the first report (16) that the "b" allele, the bT haplotype, and the bAT haplotype were preferentially transmitted to affected offspring are not supported by the data if the more informative data set is used and adjustment for multiple comparisons is made.

|
DISCUSSION |
In this meta-analysis of data from 19 reports on a possible genetic association between type 1 diabetes and four well-characterized VDR polymorphisms, we found little, if any, evidence for such an association in either case-control-type studies or family-transmission-type studies. The 95 percent confidence intervals for the pooled estimates of odds ratios or transmission proportions were fairly narrow, suggesting that large genetic effects due to these polymorphisms are unlikely. With the exception of FokI, the evidence of no association appears to be fairly consistent, as seen by the small amount of heterogeneity among pooled studies. For FokI, two out of five studies with positive findings were from the Japanese population (Figure 1), indicating that ethnicity might be a possible source of heterogeneity. Unfortunately, the small sample size and our having only two Asian population studies among 11 precluded us from conducting a meaningful investigation of possible ethnic effects. Findings from several reports that a particular polymorphism or haplotype was associated with the risk of developing type 1 diabetes, when examined more closely, turned out to be artifacts due to failure to control for multiple comparisons.
Aside from the small amount of evidence for an association after more appropriate data analysis, perhaps a more disquieting observation is the enormous inconsistency among different studies, not just between positive and negative studies but also among the positive studies. For example, while the first report by McDermott et al. (16) stated that the bAT haplotype is the high-risk haplotype, Pani et al. (24) reported that BAt is the high-risk haplotype. Again, ethnicity may be involved. However, two studies conducted in Caucasians also reported different results: One found that the combined genotype BBAAtt confers increased risk (29), and the other reported that BbAaTT and BbAATt are the high-risk genotypes and that BbAaTt is protective (33).
After pooling data from all of the published studies on the VDR-type 1 diabetes association, the fact that we found no evidence for such an association is not entirely surprising. Inconsistency aside, it seemed that the data presented in the first report (16), when analyzed more appropriately, did not support the claim of such an association. The funnel plots appeared to buttress this view: As more and more studies were conducted, the odds ratio estimate gravitated toward the null value. That having been said, we admit that we reanalyzed data on only the four most-studied SNPs in the VDR gene, and there are over 200 other SNPs in the same gene which might be associated with type 1 diabetes. Even if none of these SNPs is associated with type 1 diabetes, it is possible that VDR, as a gene mediating the action of vitamin D, may still be involved in the pathogenesis of type 1 diabetes.
The biochemical evidence for a putative relation between VDR polymorphisms and type 1 diabetes risk is very scanty. Granted, the action of vitamin D is mediated through VDR, and certain polymorphisms in the VDR gene, especially promoter or 3'-UTR (untranslated region) polymorphisms, may influence receptor transduction efficiency, transcription, or mRNA stability (13). However, it is well known that vitamin D status varies worldwide with season and not solely with diet. Without measuring an individual's vitamin D biosynthesis, it is perhaps not adequate to examine the relation between the VDR polymorphism and type 1 diabetes. This view appears to be supported by a surprising finding in VDR knockout (45) mice: They were protected from low-dose streptozotocin-induced diabetes, since immune defects observed in VDR knockout mice can be restored by calcium homeostasis normalization (46). The intriguing correlation, reported in several epidemiologic studies, between geographic latitude and the incidence of type 1 diabetes—showing an inverse correlation between monthly hours of sunshine and diabetes incidence (5)—does not always hold true: In Finland and Sardinia, which have the highest incidence of type 1 diabetes in the world, no such correlation was found (47).
The findings of this analysis appear to be consistent with a well-recognized problem in genetic association studies of complex diseases (48, 49): lack of replication. In an extensive review of 166 gene-disease associations, Hirschhorn et al. (50) found that only six (<4 percent) of the associations had been consistently replicated. Ioannidis et al. (51) reported a slightly better percentage of 16. In fact, quantitative analyses of published association studies have revealed that significant heterogeneity or lack of consistency is common, and that a small sample size in the first publication and a large number of studies are two independent predictors of discrepancies (52). In addition, the magnitude of the genetic effect differs significantly between large studies and small studies (51), as was also evidenced in this analysis. With these well-documented problems, it is perhaps prudent to exercise caution when reviewing association studies.

|
ACKNOWLEDGEMENTS |
The authors gratefully acknowledge funding from the Children's Hospital of Wisconsin Foundation and the Juvenile Diabetes Research Foundation.
The authors thank Dr. Ichiro Yokota for providing additional data and Ernest Luczkowski for retrieving literature.

|
TABLES AND FIGURES |
|

|
REFERENCES |
List of References

|
|
|
Provides link to non-governmental sites and does not necessarily represent the views of the Centers for Disease Control and Prevention. |
|
|
Page last reviewed: October 12, 2006 (archived document)
Page last updated: November 2, 2007
Content Source: National Office of Public Health Genomics |
|


