Abstract
Context Many studies try to probe for differences in risks between men and women, and this is a major challenge in the expanding literature of associations between genetic variants and common diseases or traits.
Objective To evaluate whether prominently claimed sex differences for genetic effects have sufficient internal and external validity.
Data Sources We searched PubMed through July 6, 2007, for genetic association studies claiming sex-related differences in the articles' titles. Titles and abstracts and, if necessary, the full text of the article were assessed for eligibility.
Study Selection Two hundred fifteen articles were retrieved by the search. We considered eligible all retrieved association studies that claimed different genetic effects across sexes of 1 or more gene variants for any human disease or phenotype. We considered both biallelic and multiallelic markers (including haplotypes) and both binary and continuous phenotypes and traits. We excluded non–English-language studies; studies evaluating only 1 sex; studies in which sex was treated only as an independent predictor of disease; studies that did not address any association of the investigated genetic variant with a disease or trait; studies not involving humans; and studies in which the authors did not claim any sex difference.
Data Extraction Two evaluators independently extracted data with a third evaluator arbitrating their discrepancies. Data evaluation included whether analyses were stated to have been specified a priori; whether sex effects were evaluated in the whole study or subgroups thereof; and whether the claims were appropriately documented, insufficiently documented, or spurious. For appropriately and insufficiently documented claims we performed the calculations for gene-sex interaction whenever raw data were available. Finally, we compared the sex-difference claims with the best internal validity against the results of other studies addressing the same interaction.
Results We appraised 432 sex-difference claims in 77 eligible articles. Authors stated that sex comparisons were decided a priori for 286 claims (66.2%), while the entire sample size was used in 210 (48.6%) claims. Appropriate documentation of gene-sex interaction was recorded in 55 claims (12.7%); documentation was insufficient for 303 claims and spurious for the other 74. Data for reanalysis of claims were available for 188 comparisons. Of these, 83 (44.1%) were nominally statistically significant at a P = .05 threshold, and more than half of them (n = 44) had modest P values between .01 and .05. Of 60 claims with seemingly the best internal validity, only 1 was consistently replicated in at least 2 other studies.
Conclusion In this sample of highly prominent claims of sex-related differences in genetic associations, most claims were insufficiently documented or spurious, and claims with documented good internal and external validity were uncommon.
Introduction
Sex is a factor that has been invoked extensively in the past as a modulator of effects in clinical research. However, empirical data from randomized trials suggest that many claimed subgroup differences based on sex have been spurious and led to serious misconceptions.(1) For example, aspirin was believed to be ineffective in secondary prevention of stroke in women for more than 10 years based on an underpowered subgroup analysis.(2)
In the human genome era, for many common diseases, published research has often considered that some common gene variants may have different effects in men vs women. Many diseases or traits with strong genetic backgrounds have different prevalence in the 2 sexes. For example, autoimmune diseases, endocrinopathies, and longevity are more common in women, while coronary artery disease, ischemic stroke, and high cholesterol levels are more common in men.(3) These observations do not necessarily mean that a specific gene variant should also have a different effect in men vs women. For most phenotypes, many common gene variants are likely to be responsible for determining susceptibility to disease.(4) Among autosomal variants, only some of them, if any, may interact with sex. However, given that sex information is always readily available in such studies, it is easy to test whether it influences genetic effects. Eventually, a large number of claims are made for sex differences. However, are these claims justified and valid?
Herein, we describe empirically a large sample of prominently claimed sex differences for genetic effects. We evaluated whether these claims were methodologically robust or were made based on selected and/or suboptimal analyses and with insufficient or spurious documentation. We also examined whether claims that seemingly had optimal internal validity had been corroborated by any additional studies.
Methods
Selection of Studies
We aimed to assemble a sample of studies that claimed sex subgroup differences in gene-disease associations and in which the claim was so prominent that it was mentioned even in the title of the article. Sex subgroup differences are a very common theme in the epidemiological literature. Assembling all of them or even a systematic fraction thereof would be prohibitive. Any electronic search would yield only a modest sample of the thousands of articles evaluating sex subgroup differences, since these are often listed as secondary or tertiary results. Conversely, by focusing on the title, we would favor the selection of most prominent perceived subgroup differences between sexes. These studies are the ones in which the authors (and apparently also the peer reviewers and editors) are most confident of the strength of the observed sex subgroup differences.
We searched PubMed through July 6, 2007. PubMed is considered highly representative and inclusive of genetic epidemiology studies.(5) We used a search strategy that would have high specificity for assembling a convenient sample of eligible studies: polymorphism* [ti] AND (gender [ti] OR sex [ti]). We perused titles and abstracts and, if in doubt, also the full text, for eligibility. We considered eligible all retrieved association studies that claimed different genetic effects across sexes of 1 or more gene variants for any human disease or phenotype. We considered both biallelic and multiallelic markers (including haplotypes) and both binary and continuous phenotypes and traits.
We excluded non–English-language studies; studies evaluating only 1 sex; studies in which sex was treated only as an independent predictor of disease; studies that did not address any association of the investigated genetic variant with a disease or trait; studies not involving humans; and studies in which the authors did not claim any sex difference. Eligible studies were included regardless of the extent of the quantitative information that they provided to support their claims. Eligibility assessment was performed by 2 independent evaluators (N.A.P. and A.T.). Discrepancies were resolved by consensus.
Data Extraction
Two evaluators independently extracted data, with a third evaluator arbitrating their discrepancies. We extracted the following data from each eligible study: first author, journal of publication, year, total sample size, percentage of women, gene(s) and variant(s) for which sex differences were claimed, and disease/phenotype(s) thereof.
For each pair of gene variant and phenotype for which the investigators claimed a sex difference, we recorded the exact phrasing of the claim and any allusion that the difference was tested based on a priori plans or as part of post hoc analyses; the type of genetic variant (eg, biallelic, multiallelic single-locus, haplotypes of many variants); and the type of phenotype (ie, binary outcome or continuous trait). Especially for biallelic variants, we recorded if a specific genetic contrast (recessive, dominant, additive, allele-based, model-free, or other/unclear) was used. We also recorded (per sex) the presented effect sizes and measures of uncertainty thereof or raw data that could be used to verify the presence of each claimed sex difference.
For each claim of sex difference, we recorded whether it was based on an analysis of the entire study sample or a subset thereof. If the latter, we recorded the definition of the subset and whether any a priori justification was provided for the subset selection. We specified whether subsets were defined based on genetic information for the gene variant of interest (eg, comparison of AA vs aa homozygotes without consideration of Aa heterozygotes; comparison of haplotypes 1 vs 4 without consideration of haplotypes 2 and 3), genetic information for some other gene variant (eg, selection of individuals who have some specific genotype for another gene marker), other patient characteristics (eg, age, ethnic or racial descent), other considerations, or combinations of the above.
For each claim of sex difference, we also recorded whether it was appropriately documented, insufficiently documented, or spurious. For appropriately documented claims, 3 criteria were required. First, the article had to address a genetic effect that was based on the same genetic contrast in both sexes. Second, it did not compare different subsets in the 2 sexes (eg, old men vs young women). Third, it needed to either report a nominally statistically significant (defined as a P value threshold of .05) test that examined sex-gene interaction or the interaction had to be very obvious because the presented confidence intervals of effects for each sex were not overlapping. Insufficiently documented claims were those in which only the first 2 criteria were fulfilled. Spurious claims failed either one or both of the first 2 criteria.
Insufficient documentation does not mean that the claim is necessarily inappropriate and wrong. An insufficiently documented claim may be correct (a significant interaction may exist after all) or wrong (a significant interaction may not exist), but this is not clear from the analyses performed or the way the claim is made in the article.
Reanalysis of Sex Claims
For each sex claim that was not spurious and where suitable information was available, we tried to perform calculations to test whether there was indeed a nominally statistically significant sex subgroup difference in the effect sizes (sex-gene interaction).
Whenever a dominant, recessive, or allele-based model was implied, we used for each sex the natural logarithm of the odds ratio (binary outcomes) or the absolute difference (continuous outcomes) and their variances. Whenever effect sizes were not reported for both sexes, we tried to estimate them from the presented information on published 2 x 2 tables and means per sex. Whenever variances were not reported, we tried to estimate them from the presented information on confidence intervals, standard deviation, or standard error of the mean and number of observations. We then calculated the z score as a ratio where the nominator is the difference of normalized effects and the denominator is the square root of the variance of the difference. Whenever an additive model was implied, we fitted trend models for each sex and used the resulting coefficients and respective standard errors to calculate the z score. Finally, whenever a model-free approach was implied, we used an analysis of variance with a sex-genotype interaction term.
All these analyses were performed using the data that pertain to the same subjects for which the sex subgroup difference was originally claimed by the authors. Thus, if the sex claim had been made for the entire study population, we examined sex-gene interaction in the entire population; if the claim had been made for a subset, we examined sex-gene interaction in that same subset. When both unadjusted and adjusted estimates of effect were available in full detail, we used the unadjusted estimates, except when both unadjusted and adjusted estimates were reported as different claims. In this case, both were used if possible.
Corroboration of Statistically Significant Sex-Gene Interactions by Other Studies
Even if a sex-gene interaction has been presented with optimal statistical and analytical support, it is not certain that it truly exists. Genetic effects are subject to an extensive multiplicity of testing and are also susceptible to diverse errors and biases.(6) Therefore, replication by additional independent studies is considered essential for reinforcing the credibility of genetic effects.(7) We examined whether proposed sex-gene interactions in our sample had indeed been evaluated by additional studies and, if so, whether the results of these studies agreed with the proposed interactions.
We focused on claims of sex differences that met all of the following criteria: their analyses were stated to have relied on a priori considerations, raw data were provided, sex-gene interaction was nominally statistically significant at the P = .05 level on our reanalysis of the data, and either the whole study sample had been analyzed or a subset had been selected based on a priori considerations. These claims were the ones that apparently had the best possible internal validity.
We perused in detail each of the articles reporting such claims and recorded whether they had cited any previous studies investigating the same sex-gene interaction. Additionally, we searched the ISI Web of Knowledge for articles that cited the articles meeting the criteria mentioned above. We then retrieved these cited and citing studies and examined whether they had evaluated the same sex-gene interaction and, if so, what was found.
We acknowledge that our search strategy may not have been 100% sensitive to find all studies that tested these interactions. However, there is no documented reliable strategy to retrieve all such articles, and interactions are probably often buried in text or not reported at all, especially if not nominally significant (P>.05) and "noteworthy." However, prior supporting studies are very likely to have been cited in these articles when the interactions were the main theme and even were part of the article's title. Similarly, subsequent studies that did find the same interaction would be likely to cite an article in which the main theme had been that same interaction. Overall, our strategy probably favors the retrieval of studies that agreed rather than disagreed with the proposed interactions.
Analyses were conducted in Intercooled Stata, version 8.2 (Stata Corp, College Station, Texas). P values are 2-tailed.
Results
The electronic search yielded 215 citations; 138 were excluded on close scrutiny: 5 were non–English-language papers, 34 studies evaluated only 1 sex, 11 used sex as an independent predictor, 28 were not association studies, 54 were nonhuman studies, 4 had no claim of sex difference, and we could not find the full articles for 2 citations. Seventy-seven articles (8-84) were eligible and contained a total of 432 distinct claims of sex subgroup differences (median = 4 [interquartile range {IQR}, 2-7] claims per article).
These studies were published between 1994 and 2007 in 63 different peer-reviewed journals with a median impact factor of 3.868 (IQR, 2.826-5.699). The median sample size of these studies was 560 (IQR, 274-921) and the median proportion of women was 49% (IQR, 44%-53%). Sixty-three different genes were implicated across the 432 claims. The claims pertained to a wide variety of diseases and phenotypes, the most common being hepatitis C virus infection (n = 32 claims), high-density lipoprotein cholesterol (n = 26), lung cancer (n = 21), type 2 diabetes mellitus (n = 19), multiple sclerosis (n = 18), hypertension (n = 14), low-density lipoprotein cholesterol (n = 13), Paget disease of bone (n = 12), and diabetic nephropathy in type 1 diabetes (n = 10) (Table 1).
|
TABLE 1: Evaluated Articles Making Prominent Claims for Sex Differences in Their Titles |
Reported A Priori Evaluation of Sex Claims
Of the 432 claims, 286 (66.2%) were reported as being based on a priori stated comparison of the sexes and 68 (15.7%) were acknowledged to be post hoc analyses; in the other 78 (18.1%), the analysis plan was unclear.
Genetic Variants and Phenotypes and Genetic Contrasts Involved
The variant of interest was of a biallelic locus in 328 claims, a multiallelic single locus in 44, multilocus haplotypes in 45, and multiple polymorphisms without haplotype construction in 15. The phenotype was binary in 212 claims, continuous in 218, and categorical in another 2 (cross-tabulations and other supplementary data are available at http://www.dhe.med.uoi.gr/sup_mat.php).

Among the biallelic variants, a specific genetic contrast was used in 227 claims (recessive in 33, dominant in 95, additive in 19, allele-based in 16, and model-free [all 3 genotypes considered separately] in 64). The remaining 101 were based on other, more peculiar genetic contrasts (wild-type homozygous vs variant homozygous, n = 30; heterozygous vs variant homozygous, n = 8; heterozygous vs wild-type homozygous, n = 13; and other considerations, n = 50).
Analyzed Population in Sex Claims
The entire sample size was used in 210 claims (48.6%) and the other 222 used subsets of the study population. The subset selection was based on genetic information for the gene variant of interest in 60 claims, on genetic information for some other gene variant in 2, on other patient characteristics in 101, and on combinations of the above in 59. The selection of the subset was reported to have been determined a priori in 98 claims (44.1%), it was acknowledged to have been done post hoc in 43 claims (19.4%), and the timing of the selection was unclear in 81 claims (36.5%).
Documentation of Sex Claims
Appropriate documentation was evident in only 55 claims (12.7%). This includes 49 claims that performed statistical testing for interaction and another 6 for which no formal testing was shown, but the sex-specific 95% confidence intervals of the genetic effects were readily presented and did not overlap. Forty-four (44/49 [89.8%]) of the provided P values were between .01 and .05. The smallest P value reported was .00008.
A total of 303 claims had insufficient documentation. In 81 claims, the investigators said that a statistically significant effect was found in one sex but not in the other, and both effects were in the same direction. In 46 claims, they stated that there was a statistically significant effect in one sex but not in the other, and the point estimates were in opposite directions. In 16 claims, they stated that a larger or more statistically significant sex effect was seen in one sex than in the other, and the effects were nominally statistically significant in both sexes. In 107 claims, a statistically significant effect was shown in one sex, but there was no information on statistical significance in the other sex. Finally, 53 claims reported a statistically significant effect in one sex and no statistically significant effect in the other, but the direction was not specified in either sex. For all of these situations, the way the claim was made cannot ensure whether sex-gene interaction is nominally statistically significant or not. Illustrative examples are shown in Table 2. Among the claims with insufficient documentation, 9 also reported some formal gene-sex interaction testing that was nevertheless not statistically significant at the P = .05 level (P value range, .088-.983).
|
TABLE 2: Examples of Insufficiently Documented Claims |
A total of 74 claims were spurious. The reasons for spurious claims are shown in Table 3. The most common reasons were the comparison of male vs female cases with a selected genotype, ignoring other genotypes (ie, no genetic contrast; n = 28 claims), and the comparison of male cases directly with female cases, ignoring controls (n = 8 claims). A wide variety of other comparisons were also invoked (Table 3).
|
TABLE 3: Examples of Spurious Claims |
Reanalyses of Sex-Difference Claims
Data were available for the reanalysis of 188 claims (30 appropriately documented and 158 insufficiently documented). Overall, 105 of the 188 claims (55.9%) were not nominally statistically significant based on our calculations. Illustratively, the effect sizes and 95% confidence intervals are shown in pairs for male and female participants for the non–statistically significant claims for which the effect sizes are odds ratios (n = 44) (Figure 1 and Figure 2). Eighty-three of the 188 reanalyzed claims were nominally statistically significant, but the majority (n = 44) had modest P values between .01 and .05, 25 had P values between .001 and .01, and only 14 had P values less than .001 for the interaction.
In 30 claims, the original investigators had provided appropriate statistical documentation and raw data were also available for us to retest gene-sex interaction. Of these, in 23 claims the sex difference was statistically significant in both the reported and calculated P values, while in the other 7 we could not replicate the alluded statistical significance. Six of these used multivariate analyses, while we could calculate only unadjusted estimates based on given raw data. In the remaining claim, the authors reported to have used a Breslow-Day test of homogeneity with P = .05. Our reanalysis yielded P = .059, while our Breslow-Day recalculation gave P = .054.
Corroboration of Claims With Best Internal Validity
Of the 432 claims, only 37 claims (in 17 articles) were stated to rely on a priori considerations, had raw data available that documented that they were indeed nominally statistically significant, and had analyses performed on the whole study sample. We found any kind of corroboration history for only 3 of them. One sex difference had already been described (same trend) in 2 previous studies. For another claim, a previous study had found no effect while a subsequent one replicated the interaction. The third claim had already been described in a previous study but in the opposite direction; however, 2 previous studies reported no effect (Table 4).
|
TABLE 4: Corroboration History for the Gene-Sex Interaction Claims With the Seemingly Best Internal Validity
|
Comment
We have empirically evaluated observational studies claiming to have found sex-related differences in genetic effects for common diseases and traits. Claims covered a variety of genes and outcomes. Most authors stated that these analyses had been conceived a priori. Nevertheless, the majority of these claims were insufficiently documented or spurious, and reporting of statistical interaction tests was rare. When we reanalyzed the available data, more than half of the tested gene-sex interactions failed to reach nominal statistical significance at the P = .05 level, and most of those that did reach significance had very modest P values. Even among the claims that seemingly had the best internal validity, corroboration in other studies was very rare.
Subgroup comparisons have been evaluated previously, mostly in the clinical trials literature.(93-95) To our knowledge, no such assessment exists for genetic epidemiology, although genetic determinants of common traits represent an exploding clinical research literature.(5) In the clinical trials literature, subgroup analyses with sex or other variables have been a common strategy used to find and report statistically significant results. Some authors have argued for the need to perform and transparently report subgroup analyses, in particular those related to sex.(96) However, the vast majority of claimed subgroup differences are likely to be chance findings.(97-98) Notorious chance findings were illustrated years ago in the analysis of the ISIS-2 results based on the signs of the zodiac,(99) and the principle has been further elaborated recently in simulations of subgroup analyses that highlight the major threat of false-positive results in such endeavors.(100-102) In the analysis of sex-specific effects in genetic associations, investigators very often seem to fall into classic traps. The most typical error is to claim nominal statistical significance for 1 of the 2 sexes when the difference in the effects between the 2 sexes is not beyond chance. Simulations show that a significant effect in 1 subgroup only may be a very common occurrence even in studies of modest sample size, occurring in 7% to 64% of analyses in 1 simulation study.(100-101) The majority of such claims are expected to be false-positive results.
Another major problem is the lack of power to detect interactions of effects with sex in these studies. Brookes et al have estimated that a study with 80% power for the overall effect has only 29% power to detect an interaction effect of the same magnitude.(101) Meaningful pursuit of interactions may require almost a 10-fold increase in the sample size compared with the samples needed to document main effects. (100, 103-104) At the same time, these genetic association studies are already more than 10-fold smaller than what would be required to pursue even main effects, based on what is known of the plausible size of effects for common genetic
variants. (105) Most of the main effects proposed in the last decade have not been replicated.(106) Under these circumstances, a nominally statistically significant interaction test at the P = .05 threshold probably has very low positive predictive value for the presence of a true interaction.
Some limitations should be addressed. We used a sampling strategy that was systematic but was also heavily driven by convenience. The sampling tends to select more prominently claimed sex differences, and one may assume that these are likely to have better-than-average internal validity and better-than-average chances of external corroboration. However, this cannot be formally proven. Sex is a patient characteristic available in virtually all genetic association studies, so our sample of articles is probably only the "tip of the iceberg." It is impractical to find and evaluate all sex comparisons, even in a circumscribed sample of the literature. Selective reporting of subgroup and secondary analyses is an increasingly recognized bias that may lead to a preponderance of "positive" findings in the published literature.(107-109) At a minimum, the studies that we evaluated are probably among the ones in which authors were most certain about some, if not all, of the sex claims that they presented in their results; otherwise, they would not have drawn attention to the claims in the titles of their articles.
We should also acknowledge that for some claims, especially the ones that were first made most recently, corroboration may not yet have been performed but may be performed in the future. However, genetic epidemiology is a quickly moving field, and replication efforts are currently typically performed very rapidly.
The issues addressed herein focus on gene-sex interaction, but their implications probably extend to any kind of subgroup analysis in genetics. Genetic epidemiology is a field that often invites subgroup analyses, not only by sex, but also by age, racial/ethnic descent, other polymorphisms, diet, lifestyle, and other exposures.(110-111) Similar caution may be needed in the analysis, reporting, and interpretation of all of these postulated effect modifications.
We hope that our empirical evaluation will help sensitize clinicians, geneticists, epidemiologists, and statisticians who are pursuing subgroup analyses by sex or other subgroups on genetic associations. The pursuit of gene-sex interactions should not be necessarily abandoned. Ideally, sex differences should be based on a priori, clearly defined, and adequately powered subgroups. Post hoc, discovery-based analyses are also of interest, but their post hoc character should be clearly stated in the manuscript. Both a priori and post hoc claims should be documented by interaction tests and proper consideration of the multiplicity of comparisons involved. Even then, results should be explained with caution and should be replicated by several other studies before being accepted as likely modifications of genetic or other risks.
Author Information
Corresponding Author: John P. A. Ioannidis, MD, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, University Campus, Ioannina, 45110 Greece (jioannid@cc.uoi.gr
).
Author Contributions: Dr Ioannidis had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Patsopoulos, Tatsioni, Ioannidis.
Acquisition of data: Patsopoulos, Tatsioni.
Analysis and interpretation of data: Patsopoulos, Tatsioni, Ioannidis.
Drafting of the manuscript: Patsopoulos, Ioannidis.
Critical revision of the manuscript for important intellectual content: Patsopoulos, Tatsioni, Ioannidis.
Statistical analysis: Patsopoulos, Tatsioni, Ioannidis.
Obtained funding: Ioannidis.
Study supervision: Ioannidis.
Financial Disclosures: None reported.
Funding/Support: Dr Patsopoulos was supported in part by a PENED grant sponsored by the EU European Social Fund and the Greek Ministry of Development–General Secretariat for Research and Technology.
Role of the Sponsor: The sponsor had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
Author Affiliations: Clinical and Molecular Epidemiology Unit, Department of Hygiene and Epidemiology, University of Ioannina School of Medicine (Drs Patsopoulos, Tatsioni, Ioannidis), and Biomedical Research Institute, Foundation for Research and Technology-Hellas, Ioannina, Greece (Dr Ioannidis); and Department of Medicine, Tufts University School of Medicine, Boston, Massachusetts (Drs Tatsioni and Ioannidis).
References
- Rothwell PM. Treating individuals 2: subgroup analysis in randomised controlled trials: importance, indications, and interpretation. Lancet. 2005;365(9454):176-186.
- Canadian Cooperative Study Group. A randomized trial of aspirin and sulfinpyrazone in threatened stroke. N Engl J Med. 1978;299(2):53-59.
- Harrison TR, Kasper D, Braunwald E, et al. Harrison's Principles of Internal Medicine. 16th ed. New York, NY: McGraw-Hill Education; 2004.
- Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol. 2005;34(5):1129-1137.
- Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am J Epidemiol. 2006;164(1):1-4.
- Ioannidis JP. Genetic associations: false or true? Trends Mol Med. 2003;9(4):135-138.
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306-309.
- Hansen PS, Klausen IC, Lemming L, Gerdes LU, Gregersen N, Faergeman O. Apolipoprotein B gene polymorphisms in ischemic heart disease and hypercholesterolemia: effects of age and sex. Clin Genet. 1994;45(2):78-83.
- Méplan C, Crosley LK, Nicol F, et al. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study) [published online ahead of print May 29, 2007]. FASEB J doi:10.1096/fj.07-8166com.
- Hayashi K, Maeda S, Iemitsu M, et al. Sex differences in the relationship between estrogen receptor alpha gene polymorphisms and arterial stiffness in older humans. Am J Hypertens. 2007;20(6):650-656.
- Baud P, Courtet P, Perroud N, Jollant F, Buresi C, Malafosse A. Catechol-O-methyltransferase polymorphism (COMT) in suicide attempters: a possible gender effect on anger traits [published online ahead of print May 17, 2007]. Am J Med Genet B Neuropsychiatr Genet. doi:10.1002/ajmg.b.30541.
- Yang KD, Ou CY, Hsu TY, et al. Interaction of maternal atopy, CTLA-4 gene polymorphism and gender on antenatal immunoglobulin E production. Clin Exp Allergy. 2007;37(5):680-687.
- Glorioso N, Herrera VL, Bagamasbad P, et al. Association of ATP1A1 and dear single-nucleotide polymorphism haplotypes with essential hypertension: sex-specific and haplotype-specific effects. Circ Res. 2007;100(10):1522-1529.
- Gallagher CJ, Muscat JE, Hicks AN, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(4):823-828.
- Beyens G, Daroszewska A, de Freitas F, et al. Identification of sex-specific associations between polymorphisms of the osteoprotegerin gene, TNFRSF11B, and Paget's disease of bone. J Bone Miner Res. 2007;22(7):1062-1071.
- Leung KH, Yip SP, Wong WS, et al. Sex- and age-dependent association of SLC11A1 polymorphisms with tuberculosis in Chinese: a case control study. BMC Infect Dis. 2007;7:19.
- Bolufer P, Collado M, Barragán E, et al. The potential effect of gender in combination with common genetic polymorphisms of drug-metabolizing enzymes on the risk of developing acute leukemia. Haematologica. 2007;92(3):308-314.
- Yamaguchi S, Yamada Y, Matsuo H, et al. Gender differences in the association of gene polymorphisms with type 2 diabetes mellitus. Int J Mol Med. 2007;19(4):631-637.
- Russo P, Loguercio M, Lauria F, et al, European Collaborative Group of the IMMIDIET Project. Age- and gender-dependent association of the -344C/T polymorphism of CYP11B2 with blood pressure in European populations. J Hum Hypertens. 2007;21(4):333-336.
- Froehlich TE, Lanphear BP, Dietrich KN, Cory-Slechta DA, Wang N, Kahn RS. Interactive effects of a DRD4 polymorphism, lead, and sex on executive functions in children [published online ahead of print]. Biol Psychiatry. 2007;62(3):243-249.
- Dedoussis GV, Theodoraki EV, Manios Y, et al. The Pro12Ala polymorphism in PPAR
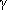 2 gene affects lipid parameters in Greek primary school children: a case of gene-to-gender interaction. Am J Med Sci. 2007;333(1):10-15. 2 gene affects lipid parameters in Greek primary school children: a case of gene-to-gender interaction. Am J Med Sci. 2007;333(1):10-15.
- Barnett JH, Heron J, Ring SM, et al. Gender-specific effects of the catechol-O-methyltransferase Val108/158Met polymorphism on cognitive function in children. Am J Psychiatry. 2007;164(1):142-149.
- Schott E, Witt H, Hinrichsen H, et al. Gender-dependent association of CTLA4 polymorphisms with resolution of hepatitis C virus infection. J Hepatol. 2007;46(3):372-380.
- Niemi M, Pasanen MK, Neuvonen PJ. SLCO1B1 polymorphism and sex affect the pharmacokinetics of pravastatin but not fluvastatin. Clin Pharmacol Ther. 2006;80(4):356-366.
- Asselbergs FW, Williams SM, Hebert PR, et al. The gender-specific role of polymorphisms from the fibrinolytic, renin-angiotensin, and bradykinin systems in determining plasma t-PA and PAI-1 levels. Thromb Haemost. 2006;96(4):471-477.
- Yiannakouris N, Melistas L, Yannakoulia M, Mungal K, Mantzoros CS. The –2548G/A polymorphism in the human leptin gene promoter region is associated with plasma free leptin levels; interaction with adiposity and gender in healthy subjects. Hormones (Athens). 2003;2(4):229-236.
- Seripa D, Franceschi M, Matera MG, et al. Sex differences in the association of apolipoprotein E and angiotensin-converting enzyme gene polymorphisms with healthy aging and longevity: a population-based study from southern Italy. J Gerontol A Biol Sci Med Sci. 2006;61(9):918-923.
- Paladino N, Fainboim H, Theiler G, et al. Gender susceptibility to chronic hepatitis C virus infection associated with interleukin 10 promoter polymorphism. J Virol. 2006;80(18):9144-9150.
- Körner A, Ma L, Franks PW, et al. Sex-specific effect of the Val1483Ile polymorphism in the fatty acid synthase gene (FAS) on body mass index and lipid profile in Caucasian children. Int J Obes (Lond). 2007;31(2):353-358.
- Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI. Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer's disease. Neurobiol Aging. 2007;28(6):856-862.
- Ozawa T, Ito M, Tamaki S, et al. Gender and age effects on ventricular repolarization abnormality in Japanese general carriers of a G643S common single nucleotide polymorphism for the KCNQ1 gene. Circ J. 2006;70(6):645-650.
- Ben Assayag E, Bova I, Berliner S, et al. Gender differences in the expression of erythrocyte aggregation in relation to B beta-fibrinogen gene polymorphisms in apparently healthy individuals. Thromb Haemost. 2006;95(3):428-433.
- Kates WR, Antshel KM, Abdulsabur N, et al. A gender-moderated effect of a functional COMT polymorphism on prefrontal brain morphology and function in velo-cardio-facial syndrome (22q11.2 deletion syndrome). Am J Med Genet B Neuropsychiatr Genet. 2006;141(3):274-280.
- Mizuno T, Aoki M, Shimada Y, et al. Gender difference in association between polymorphism of serotonin transporter gene regulatory region and anxiety. J Psychosom Res. 2006;60(1):91-97.
- Mlynarski WM, Placha GP, Wolkow PP, Bochenski JP, Warram JH, Krolewski AS. Risk of diabetic nephropathy in type 1 diabetes is associated with functional polymorphisms in RANTES receptor gene (CCR5): a sex-specific effect. Diabetes. 2005;54(11):3331-3335.
- Derzbach L, Treszl A, Balogh A, Vasarhelyi B, Tulassay T, Rigo JJ. Gender dependent association between perinatal morbidity and estrogen receptor-alpha Pvull polymorphism. J Perinat Med. 2005;33(5):461-462.
- Gloria-Bottini F, Bottini N, Cosmi E, Cosmi EV, Bottini E. The effect of gender and ACP1 genetic polymorphism on the correlation between birth weight and placental weight. Placenta. 2005;26(10):846-848.
- Sjöberg RL, Nilsson KW, Nordquist N, et al. Development of depression: sex and the interaction between environment and a promoter polymorphism of the serotonin transporter gene. Int J Neuropsychopharmacol. 2006;9(4):443-449.
- Tan EC, Chong SA, Wang H, Chew-Ping Lim E, Teo YY. Gender-specific association of insertion/deletion polymorphisms in the nogo gene and chronic schizophrenia. Brain Res Mol Brain Res. 2005;139(2):212-216.
- Shi Q, Zhang Z, Li G, et al. Sex differences in risk of lung cancer associated with methylene-tetrahydrofolate reductase polymorphisms. Cancer Epidemiol Biomarkers Prev. 2005;14(6):1477-1484.
- Foltynie T, Lewis SG, Goldberg TE, et al. The BDNF Val66Met polymorphism has a gender specific influence on planning ability in Parkinson's disease. J Neurol. 2005;252(7):833-838.
- Kajinami K, Brousseau ME, Lamon-Fava S, Ordovas JM, Schaefer EJ. Gender-specific effects of estrogen receptor alpha gene haplotype on high-density lipoprotein cholesterol response to atorvastatin: interaction with apolipoprotein AI gene polymorphism. Atherosclerosis. 2005;178(2):331-338.
- Kantarci OH, Goris A, Hebrink DD, et al. IFNG polymorphisms are associated with gender differences in susceptibility to multiple sclerosis. Genes Immun. 2005;6(2):153-161.
- Chang JC, Liu CA, Chuang H, et al. Gender-limited association of cytotoxic T-lymphocyte antigen-4 (CTLA-4) polymorphism with cord blood IgE levels. Pediatr Allergy Immunol. 2004;15(6):506-512.
- Cui J, Randell E, Renouf J, et al. Gender dependent association of thrombospondin-4 A387P polymorphism with myocardial infarction. Arterioscler Thromb Vasc Biol. 2004;24(11):e183-e184.
- Schrijver HM, Crusius JB, Garcia-Gonzalez MA, et al. Gender-related association between the TGFB1+869 polymorphism and multiple sclerosis. J Interferon Cytokine Res. 2004;24(9):536-542.
- Liu G, Zhou W, Wang LI, et al. MPO and SOD2 polymorphisms, gender, and the risk of non-small cell lung carcinoma. Cancer Lett. 2004;214(1):69-79.
- Szczeklik W, Sanak M, Szczeklik A. Functional effects and gender association of COX-2 gene polymorphism G-765C in bronchial asthma. J Allergy Clin Immunol. 2004;114(2):248-253.
- Corder EH, Ghebremedhin E, Taylor MG, Thal DR, Ohm TG, Braak H. The biphasic relationship between regional brain senile plaque and neurofibrillary tangle distributions: modification by age, sex, and APOE polymorphism. Ann N Y Acad Sci. 2004;1019:24-28.
- Chen W, Duan S, Zhou J, et al. A case-control study provides evidence of association for a functional polymorphism –197C/G in XBP1 to schizophrenia and suggests a sex-dependent effect. Biochem Biophys Res Commun. 2004;319(3):866-870.
- Kajinami K, Brousseau ME, Ordovas JM, Schaefer EJ. Polymorphisms in the multidrug resistance-1 (MDR1) gene influence the response to atorvastatin treatment in a gender-specific manner. Am J Cardiol. 2004;93(8):1046-1050.
- Yang KD, Liu CA, Chang JC, et al. Polymorphism of the immune-braking gene CTLA-4 (+49) involved in gender discrepancy of serum total IgE levels and allergic diseases. Clin Exp Allergy. 2004;34(1):32-37.
- Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest. 2004;125(1):203-211.
- Ko YL, Hsu LA, Hsu KH, Ko YH, Lee YS. The interactive effects of hepatic lipase gene promoter polymorphisms with sex and obesity on high-density-lipoprotein cholesterol levels in Taiwanese-Chinese. Atherosclerosis. 2004;172(1):135-142.
- Nakanishi S, Yamane K, Kamei N, Okubo M, Kohno N. The effect of polymorphism in the intestinal fatty acid-binding protein 2 gene on fat metabolism is associated with gender and obesity amongst non-diabetic Japanese-Americans. Diabetes Obes Metab. 2004;6(1):45-49.
- Chen Y, Vaughan RW, Kondeatis E, et al. Chemokine gene polymorphisms associate with gender in patients with uveitis. Tissue Antigens. 2004;63(1):41-45.
- Espino-Montoro A, Barrios-Artillo M, Lopez-Chozas JM, Cayuela A, Stiefel P, Villar J. Influence of polymorphism (RFLP-sstI) at the apolipoprotein C-III gene locus on the lipoprotein metabolism and insulin resistance in essential hypertensive patients: interaction between gender and genetic polymorphism. Nutr Metab Cardiovasc Dis. 2003;13(4):194-201.
- Vandenbroeck K, Cunningham S, Goris A, et al. Polymorphisms in the interferon-gamma/interleukin-26 gene region contribute to sex bias in susceptibility to rheumatoid arthritis. Arthritis Rheum. 2003;48(10):2773-2778.
- Song J, Narita I, Goto S, et al. Gender specific association of aldosterone synthase gene polymorphism with renal survival in patients with IgA nephropathy. J Med Genet. 2003;40(5):372-376.
- Laule M, Meisel C, Prauka I, et al. Interaction of CA repeat polymorphism of the endothelial nitric oxide synthase and hyperhomocysteinemia in acute coronary syndromes: evidence of gender-specific differences. J Mol Med. 2003;81(5):305-309.
- Reich H, Duncan JA, Weinstein J, Cattran DC, Scholey JW, Miller JA. Interactions between gender and the angiotensin type 1 receptor gene polymorphism. Kidney Int. 2003;63(4):1443-1449.
- Watson MA, Gay L, Stebbings WS, Speakman CT, Bingham SA, Loktionov A. Apolipoprotein E gene polymorphism and colorectal cancer: gender-specific modulation of risk and prognosis. Clin Sci (Lond). 2003;104(5):537-545.
- Stanković A, Zivkovic M, Alavantic D. Angiotensin I-converting enzyme gene polymorphism in a Serbian population: a gender-specific association with hypertension. Scand J Clin Lab Invest. 2002;62(6):469-475.
- Györffy B, Vasarhelyi B, Krikovszky D, et al. Gender-specific association of vitamin D receptor polymorphism combinations with type 1 diabetes mellitus. Eur J Endocrinol. 2002;147(6):803-808.
- Minihane AM, Finnegan YE, Talmud P, Leigh-Firbank EC, Williams CM. Influence of the APOC3 –2854T>G polymorphism on plasma lipid levels: effect of age and gender. Biochim Biophys Acta. 2002;1583(3):311-314.
- Wu Y, Tomon M, Sumino K. Methylenetetrahydrofolate reductase gene polymorphism and ischemic stroke: sex difference in Japanese. Kobe J Med Sci. 2001;47(6):255-262.
- Lio D, Scola L, Crivello A, et al. Gender-specific association between -1082 IL-10 promoter polymorphism and longevity. Genes Immun. 2002;3(1):30-33.
- Liu ZH, Cheng ZH, Gong RJ, Liu H, Liu D, Li LS. Sex differences in estrogen receptor gene polymorphism and its association with lupus nephritis in Chinese. Nephron. 2002;90(2):174-180.
- Sciacca FL, Ferri C, Veglia F, et al. IL-1 genes in myasthenia gravis: IL-1A –889 polymorphism associated with sex and age of disease onset. J Neuroimmunol. 2002;122(1-2):94-99.
- Ordovas JM, Corella D, Cupples LA, et al. Polyunsaturated fatty acids modulate the effects of the APOA1 G-A polymorphism on HDL-cholesterol concentrations in a sex-specific manner: the Framingham Study. Am J Clin Nutr. 2002;75(1):38-46.
- Oh JY, Barrett-Connor E, Rancho Bernardo Study Group. Apolipoprotein E polymorphism and lipid levels differ by gender and family history of diabetes: the Rancho Bernardo Study. Clin Genet. 2001;60(2):132-137.
- Ellsworth DL, Bielak LF, Turner ST, Sheedy PF II, Boerwinkle E, Peyser PA. Gender- and age-dependent relationships between the E-selectin S128R polymorphism and coronary artery calcification. J Mol Med. 2001;79(7):390-398.
- Du L, Bakish D, Hrdina PD. Gender differences in association between serotonin transporter gene polymorphism and personality traits. Psychiatr Genet. 2000;10(4):159-164.
- Bullido MJ, Guallar-Castillon P, Artiga MJ, et al. Alzheimer's risk associated with human apolipoprotein E, alpha-2 macroglobulin and lipoprotein receptor related protein polymorphisms: absence of genetic interactions, and modulation by gender. Neurosci Lett. 2000;289(3):213-216.
- Kark JD, Sinnreich R, Leitersdorf E, Friedlander Y, Shpitzen S, Luc G. Taq1B CETP polymorphism, plasma CETP, lipoproteins, apolipoproteins and sex differences in a Jewish population sample characterized by low HDL-cholesterol. Atherosclerosis. 2000;151(2):509-518.
- Durlach A, Clavel C, Girard-Globa A, Durlach V. Sex-dependent association of a genetic polymorphism of cholesteryl ester transfer protein with high-density lipoprotein cholesterol and macrovascular pathology in type II diabetic patients. J Clin Endocrinol Metab. 1999;84(10):3656-3659.
- Sagnella GA, Rothwell MJ, Onipinla AK, Wicks PD, Cook DG, Cappuccio FP. A population study of ethnic variations in the angiotensin-converting enzyme I/D polymorphism: relationships with gender, hypertension and impaired glucose metabolism. J Hypertens. 1999;17(5):657-664.
- Reynolds WF, Rhees J, Maciejewski D, et al. Myeloperoxidase polymorphism is associated with gender specific risk for Alzheimer's disease. Exp Neurol. 1999;155(1):31-41.
- Freire MB, Ji L, Onuma T, Orban T, Warram JH, Krolewski AS. Gender-specific association of M235T polymorphism in angiotensinogen gene and diabetic nephropathy in NIDDM. Hypertension. 1998;31(4):896-899.
- Suarez F, Zeghoud F, Rossignol C, Walrant O, Garabedian M. Association between vitamin D receptor gene polymorphism and sex-dependent growth during the first two years of life. J Clin Endocrinol Metab. 1997;82(9):2966-2970.
- Lehtimäki T, Frankberg-Lakkala H, Solakivi T, et al. The effect of short-term fasting, apolipoprotein E gene polymorphism, and sex on plasma lipids. Am J Clin Nutr. 1997;66(3):599-605.
- Carter AM, Catto AJ, Bamford JM, Grant PJ. Gender-specific associations of the fibrinogen B beta 448polymorphism, fibrinogen levels, and acute cerebrovascular disease. Arterioscler Thromb Vasc Biol. 1997;17(3):589-594.
- Ferrières J, Sing CF, Roy M, Davignon J, Lussier-Cacan S. Apolipoprotein E polymorphism and heterozygous familial hypercholesterolemia: sex-specific effects. Arterioscler Thromb. 1994;14(10):1553-1560.
- von Eckardstein A, Funke H, Chirazi A, et al. Sex-specific effects of the glutamine/histidine polymorphism in apo A-IV on HDL metabolism. Arterioscler Thromb. 1994;14(7):1114-1120.
- Higaki J, Baba S, Katsuya T, et al. Deletion allele of angiotensin-converting enzyme gene increases risk of essential hypertension in Japanese men: the Suita study. Circulation. 2000;101(17):2060-2065.
- O’Donnell CJ, Lindpaintner K, Larson MG, et al. Evidence for association and genetic linkage of the angiotensin-converting enzyme locus with hypertension and blood pressure in men but not women in the Framingham heart study. Circulation. 1998;97(18):1766-1772.
- Ahmed AR, MacGowan SH, Culpan D, Jones RW, Wilcock GK. The –491A/T polymorphism of the apolipoprotein E gene is associated with the ApoEe4 allele and Alzheimer's disease. Neurosci Lett. 1999;263(2-3):217-219.
- Parra-Bonilla G, Arboleda G, Yunis J, et al. Haplogroup analysis of the risk associated with APOE promoter polymorphisms (2219T/G, 2491A/T and 2427T/C) in Colombian Alzheimer's disease patients. Neurosci Lett. 2003;349(3):159-162.
- Meng QH, Pajukanta P, Valsta L, Aro A, Pietinen P, Tikkanen MJ. Influence of apolipoprotein A-1 promoter polymorphism on lipid levels and responses to dietary change in Finnish adults. J Intern Med. 1997;214(5):373-378.
- Mata P, Lopez-Miranda J, Pocovi M, et al. Human apolipoprotein A-I gene promoter mutation influences plasma low density lipoprotein cholesterol response to dietary fat saturation. Atherosclerosis. 1998;137(2):367-376.
- Xu CF, Angelico F, Del Ben M, Humphries S. Role of genetic variation at the Apo AI-CIII-AIV gene cluster in determining plasma Apo Al levels in boys and girls. Genet Epidemiol. 1993;10(2):113-122.
- Kotze MJ, De Villiers WJ, Steyn K, et al. Phenotypic variation among familial hypercholesterolemics heterozygous for either one of two Afrikaner founder LDL receptor mutations. Arterioscler Thromb. 1993;13(10):1460-1468.
- Assmann SF, Pocock SJ, Enos LE, Kasten LE. Subgroup analysis and other (mis)uses of baseline data in clinical trials. Lancet. 2000;355(9209):1064-1069.
- Pocock SJ, Assmann SF, Enos LE, Kasten LE. Subgroup analysis, covariate adjustment and baseline comparisons in clinical trial reporting: current practice and problems. Stat Med. 2002;21(19):2917-2930.
- Hernández AV, Steyerberg EW, Gillian ST, Marmarou A, Habbema JD, Maas AI. Subgroup analysis and covariate adjustment in randomized clinical trials of traumatic brain injury: a systematic review. Neurosurgery. 2005;57(6):1244-1253.
- Rochon PA, Clark JP, Binns MA, Patel V, Guwitz JH. Reporting of gender-related information in clinical trials of drug therapy for myocardial infraction. CMAJ. 1998;159(4):321-327.
- Oxman AD, Guyatt GH. A consumer's guide to subgroup analysis. Ann Intern Med. 1992;116(1):78-84.
- Freemantle N. Interpreting the results of secondary end points and subgroup analyses in clinical trials: should we lock the crazy aunt in the attic? BMJ. 2001;322(7292):989-991.
- Collins R, Peto R, Gray R, Paris S. Large-scale randomised evidence: trials and overviews. In: Weatherall DJ, Ledingham JGG, Warrell DA, eds. Oxford Textbook of Medicine. London, England: Oxford University Press; 1996:21-22.
- Brookes ST, Whitley E, Peters TJ, Mulheran PA, Egger M, Davey Smith G. Subgroup analyses in randomized controlled trials: quantifying the risks of false-positives and false-negatives. Health Technol Assess. 2001;5(33):1-56.
- Brookes ST, Whitely E, Egger M, Smith GD, Mulheran PA, Peters TJ. Subgroup analyses in randomized trials: risks of subgroup-specific analyses; power and sample size for the interaction test. J Clin Epidemiol. 2004;57(3):229-236.
- Austin PC, Mamdani MM, Juurlink DN, Hux JE. Testing multiple statistical hypotheses resulted in spurious associations: a study of astrological signs and health. J Clin Epidemiol. 2006;59(9):964-969.
- Yusuf S, Wittes J, Probstfield J, Tyroler HA. Analysis and interpretation of treatment effects in subgroups of patients in randomized clinical trials. JAMA. 1991;266(1):93-98.
- Greenland S. Tests for interaction in epidemiologic studies: a review and a study of power. Stat Med. 1983;2(2):243-251.
- Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164(7):609-614.
- Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297(14):1551-1561.
- Hahn S, Williamson PR, Hutton JL, Garner P, Flynn EV. Assessing the potential for bias in meta-analysis due to selective reporting of subgroup analyses within studies. Stat Med. 2000;19(24):3325-3336.
- Chan AW, Hrobjartsson A, Haahr MT, Gotzsche PC, Altman DG. Empirical evidence for selective reporting of outcomes in randomized trials: comparison of protocols to published articles. JAMA. 2004;291(20):2457-2465.
- Chan AW, Altman DG. Identifying outcome reporting bias in randomised trials on PubMed: review of publications and survey of authors. BMJ. 2005;330(7494):753.
- Ioannidis JP, Ntzani EE, Trikalinos TA. "Racial" differences in genetic effects for complex diseases. Nat Genet. 2004;36(12):1312-1318.
- Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287-298.
|


