ISSN: 1080-6059
Volume 14, Number 7–July 2008
Dispatch
New qnr Gene Cassettes Associated with Superintegron Repeats in Vibrio cholerae O1
Érica L. Fonseca,* ![]() Fernanda dos Santos Freitas,* Verônica V. Vieira,† and
Ana C.P. Vicente*
Fernanda dos Santos Freitas,* Verônica V. Vieira,† and
Ana C.P. Vicente*
*Instituto Oswaldo Cruz, Rio de Janeiro, Brazil; and †Instituto Nacional de
Controle de Qualidade em Saúde, Rio de Janeiro
Suggested citation for this article
Abstract
A novel qnr determinant emerged in ciprofloxacin-resistant Vibrio
cholerae O1 from the Amazon region of Brazil. This qnrVC1 was in a
typical class 1 integron. Its attC showed 89% identity with V.
parahaemolyticus superintegron repeats. Analysis showed V. cholerae O1 carrying qnrVC2 associated with a V. cholerae superintegron repeat.
Quinolones are antimicrobial drugs effective against gram-negative bacteria. These drugs have been widely used as alternatives to conventional drug therapy for treating Mycobacterium spp. and Neisseria spp. infections. Quinolone resistance is frequently caused by mutations in DNA gyrase and topoisomerase IV chromosomal genes and an increased activity of efflux pumps that reduce intracellular concentrations of the drug (1).
Resistance to these drugs has been attributed to plasmid-mediated qnr genes. These genes are found mainly in Enterobacteriaceae and affect the dynamics of development and acquisition of quinolone resistance. Until now, all plasmid-borne qnr determinants were associated with atypical sul-type integrons, which are characterized by a duplication of a 3′ conserved segment, and a putative recombinase known as open reading frame (ORF) 513 (2). In contrast to typical gene cassettes, these qnr genes lack the attC site and, consequently, are unable to move by class 1 integrase excision. Qnr-like proteins coded by chromosomal genes have been found in Vibrionaceae, and this family has been identified as the source of qnr genes (3,4). However, no qnr homolog was found in any Vibrio cholerae genome sequenced.
Members of the family Vibrionaceae are characterized by superintegrons (SIs), which are chromosomal genetic elements containing a variety of gene cassettes with nearly identical attC sites (e.g., V. cholerae SI repeats) (5). We report 2 new qnr-like genes (qnrVC1 and qnrVC2) in typical class 1 integrons from clinical strains of V. cholerae O1.
The Study
V. cholerae O1 isolates from the cholera epidemic in Brazil (1991–2000) were screened for antimicrobial drug resistance and class 1 integrons. VC627, a clinical strain from the Amazon region isolated in 1998, was resistant to ciprofloxacin, in contrast to other isolates. This strain was the only one positive for class 1 integrase but not for qacEΔ1 and sulI genes (6). Sequence analysis of the variable region of this class 1 integron showed an aadA2 gene cassette and an ORF similar to qnr determinants. Its deduced amino acid sequence showed 69% sequence identity with a chromosomal protein of Photobacterium profundum, which was recently identified as a quinolone-resistance determinant (QnrPP) (3) and 57% identity with QnrA1, a plasmid-encoded protein found in Enterobacteriaceae (2). This new qnr-like gene in V. cholerae was named qnrVC1 (EU436855).
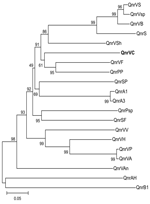 |
Figure. Genetic relationships of plasmid- and chromosome-encoded qnr proteins... |
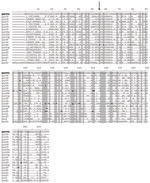 |
Appendix Figure.Deduced amino acid sequence comparison of QnrVC1 from class 1 integron with plasmid- and chromosomal-mediated qnr determinants... |
Qnr proteins belong to the pentapeptide repeat family, which consists of uninterrupted pentapeptide repeats comprising 92% of the sequence and play a role in DNA gyrase protection (7). These repeat features were observed in the QnrVC1 deduced amino acid sequence with the consensus sequence [AC][DN][LF]XX (amino acids in brackets are found more frequently and XX indicates other amino acids) (8) ( Appendix Figure), with a large proportion of serine residues in the fourth position (first X), as observed by Robicsek et al. (2).
The qnrVC homologs in chromosomes of other bacterial species were investigated by using BLAST analysis (www.ncbi.nlm.nih.gov/blast/Blast.cgi). Similar to other reports of such homologs (3,4), we found Qnr-like proteins in recently published genomes from different families, including other members of Vibrionaceae (Figure). Analysis in conjunction with informatics capabilities (in silico analysis) showed a 657-nt sequence in a class 1 integron of V. cholerae O1 isolated in Vietnam in 2004 (AB200915). This ORF had 75% nt similarity with qnrVC1, and it was designated qnrVC2.
The qnrVC gene cassettes are characterized by attC sites, which are absent in all others qnr genes already identified. Supporting the hypothesis of Vibrionaceae as the source of qnr genes (3), the qnrVC1 attC site has 89% identity with V. parahaemolyticus repeats, and the qnrVC2 putative attC site has 96% identity with a V. cholerae repeat sequence, both of which are characteristic structures of SIs. Rowe-Magnus et al. (5) reported that SI repeat recombination sites are species specific and these elements would be the source of gene cassettes found in drug-resistance integrons, such as class 1 integrons. Therefore, we hypothesize that qnrVC1 gene cassette could have originated from a V. parahaemolyticus SI, whereas qnrVC2 originated in V. cholerae. Curiously, no chromosomal qnr genes identified in Vibrionaceae were associated with SIs.
The qnrVC1 gene showed high amino acid divergence compared with all qnr genes described, with similarities ranging from 44% to 69%. The qnrVC1 had a G + C content of 36.8%, which is considerably different from that of the V. cholerae genome (47.5%) and is evidence of horizontal gene transfer.
Promoter Pc, which is found in the 5′ conserved segment of the integron carrying qnrVC1, was identical to the hybrid configuration described (9,10) and is defined by 1 point mutation in the canonical –35 region. The hybrid Pc version has 20-fold lower promoter activity than that of the wild-type (strong) promoter. However, another study verified expression of genes inserted into integrons under the control of a hybrid Pc configuration (11). Moreover, in silico analysis detected a putative promoter (–35 TTGAGA [17 nt] –10 TAGTCT) in the 5′ untranslated region of the qnrVC1 gene cassette. Therefore, our findings characterize conditions for qnrVC1 expression in the class 1 integron.
Quinolones have excellent antimicrobial activity against V. cholerae (12). Strain VC627 shows a 10-fold increase in resistance to ciprofloxacin (MIC 0.25 μg/mL) compared with strains from the seventh pandemic lineage in Brazil (MIC 0.02 μg/mL). This increased resistance was also observed in a Shigella flexneri strain harboring qnrS (13,14) and V. splendidus harboring chromosomal qnrVS1 and qnrVS2 (4). This MIC value resembles that caused by gyrA mutations in V. cholerae ciprofloxacin-resistant strains (15). Therefore, dissemination of qnr genes by lateral gene transfer may determine, in a 1-step fashion, the same drug-resistance profile caused by acquired mutations in housekeeping genes.
Conclusions
Our study reports characterization of 2 new qnr genes associated with SI attC sites in typical class 1 integrons and their emergence in V. cholerae. These genetic features have not been observed in either chromosomal- or plasmid-borne qnr determinants already described.
Our study suggests that, at least in Vibrionaceae, quinolone-resistance genes were mobilized from SIs by class 1 integrase activity because qnrVC1 and qnrVC2 attC elements are similar to V. parahaemolyticus and V. cholerae SI repeats. These findings corroborate the hypothesis that SIs are a source of cassettes present in drug-resistance integrons (5). These elements are part of a reasonable scenario for a 1-step acquisition of quinolone resistance by bacteria (e.g., Mycobacterium spp. and Neisseria spp.). This acquisition may jeopardize treatment and control and have adverse consequences on infections caused by these organisms.
Acknowledgments
This work was supported by a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico, a grant from the Instituto Oswaldo Cruz, and the Plataforma de Desenvolvimento Technólogico em Insumos para a Saúde for sequencing.
Ms Fonseca is a doctoral student at the Laboratório de Genética Molecular de Microrganismos, Instituto Oswaldo Cruz, FIOCRUZ, in Rio de Janeiro. Her research interest is genetic variability of microorganisms, especially bacteria of clinical relevance.
References
- Hawkey PM. Mechanisms of quinolone action and microbial response. J Antimicrob Chemother. 2003;51:29–35. PubMed DOI
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40. PubMed DOI
- Poirel L, Liard A, Rodriguez-Martinez J-M, Nordmann P. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J Antimicrob Chemother. 2005;56:1118–21. PubMed DOI
- Cattoir V, Poirel L, Mazel D, Soussy C-J, Nordmann P. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob Agents Chemother. 2007;51:2650–1. PubMed DOI
- . Rowe-Magnus DA, Guérout A-M, Ploncard P, Dychinco B, Davies J, Mazel D. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc Natl Acad Sci U S A. 2001;98:652–7. PubMed DOI
- Fonseca EL, Vieira VV, Cipriano R, Vicente AC. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol. 2005;44:303–9. PubMed DOI
- Vetting MW, Hegde SS, Fajardo JE, Fiser A, Roderick SL, Takiff HE, et al. Pentapeptide repeat proteins. Biochemistry. 2006;45:1–10. PubMed DOI
- Tran JH, Jacoby GA. Mechanism of plasmid-mediated quinolone resistance. Proc Natl Acad Sci U S A. 2002;99:5638–42. PubMed DOI
- Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62.
- Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. PubMed DOI
- Dubois V, Poirel L, Marie C, Arpin C, Nordmann P, Quentin C. Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2002;46:638–45. PubMed DOI
- Yamamoto T, Nair GB, Albert MJ, Parodi CC, Takeda Y. Survey of in vitro susceptibilities of Vibrio cholerae O1 and O139 to antimicrobial agents. Antimicrob Agents Chemother. 1995;39:241–4.
- Hata M, Suzuki M, Matsumoto M, Takahashi M, Sato K, Ibe S, et al. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob Agents Chemother. 2005;49:801–3. PubMed DOI
- Martínez-Martínez L, Pascual A, García I, Tran J, Jacoby GA. Interaction of plasmid and host quinolone resistance. J Antimicrob Chemother. 2003;51:1037–9. PubMed DOI
- Baranwal S, Dey K, Ramamurthy T, Nair GB, Kundu M. Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob Agents Chemother. 2002;46:2676–8. PubMed DOI
Figures
Figure. Genetic relationships of plasmid- and chromosome-encoded qnr proteins...
Appendix Figure. Deduced amino acid sequence comparison of QnrVC1 from class 1 integron with plasmid- and chromosomal-mediated qnr determinants...
Suggested Citation for this Article
Fonseca EL, Freitas FS, Vieira VV, Vicente ACP. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg Infect Dis [serial on the
Internet]. 2008 Jul [date cited]. Available from
http://www.cdc.gov/EID/content/14/7/1129.htm
DOI:10.3201/eid1407.080132
Please use the form below to submit correspondence to the authors or contact them at the following address:
Érica L. Fonseca, Laboratório de Genética Molecular de Microrganismos, Pavilhão Leônidas Deane, Sala 607, Instituto Oswaldo Cruz, FIOCRUZ, Avenida Brazil 4365, PO Box 926, Manguinhos, Rio de Janeiro, Brazil; email: ericafon@ioc.fiocruz.br
Please contact the EID Editors at eideditor@cdc.gov
The opinions expressed by authors contributing to this journal do not necessarily reflect the opinions of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
This page posted June 30, 2008
Centers for Disease Control and Prevention, 1600 Clifton Rd, Atlanta, GA 30333, U.S.A
Tel: (404) 639-3311 / Public Inquiries: (404) 639-3534 / (800) 311-3435