Reference
Complete the bibliographic reference for the article according to AJE format.
|
Stephens JW et al. The D allele of the ACE I/D common gene variant is associated with Type 2 diabetes mellitus in Caucasian subjects. Mol Genet Metab 2005;84:83-9. |
Category of HuGE information
Specify the types of information (from the list below) available in the article:
- Prevalence of gene variant
- Gene-disease association
- Gene-environment interaction
- Gene-gene interaction
- Genetic test evaluation/monitoring
|
2. Gene-disease association |
Study hypotheses or purpose
The authors study hypotheses or main purpose for conducting the study |
The main goal of the paper was to compare the allele frequency and genotype distribution of the angiotensin-1 converting enzyme (ACE) insertion (I)/deletion (D) polymorphism in Caucasians with and without Type 2 diabetes mellitus (DM).
|
Gene(s)
Identification of the following:
- Gene name
- Chromosome location
- Gene product/function
- Alleles
- OMIM #
- GDPInfo link
|
- Gene name: Angiotensin-1 converting enzyme (ACE)
- Chromosome location: 17q23
- Gene product/function: ACE is a dipeptidyl carboxypeptidase that plays an important role in blood pressure regulation and electrolyte balance by hydrolyzing angiotensin I into angiotensin II and aldosterone-stimulating peptide. Angiotensin II stimulates vasoconstriction and sodium retention. ACE is also able to inactivate bradykinin, a potent vasodilator. Thus, ACE acts to increase blood pressure. Angiotensin II is also pro-inflammatory and pro-oxidant and thus can cause cellular toxicity and apoptosis, while bradykinin may enhance the activity of insulin. Thus increased ACE activity may promote inflammation and the development of Type 2 Diabetes.
- Alleles: Insertion (I)/Deletion (D) polymorphism (ACE I/D). The I allele has a 287 bp insertion in intron 16. The D allele leads to higher ACE expression and activity in Caucasians. In Caucasians, the I/D variant accounts for about 50% of the phenotypic variance in serum ACE.
- OMIM #: 106180
- Go to GDPInfo Genes A-Z result
|
Environmental factor(s)
Identification of the major environmental factors studied (infectious, chemical, physical, nutritional, and behavioral) |
None |
Health outcome(s)
Identification of the major health outcome(s) studied
|
- Type 2 diabetes mellitus
- Family history of Type 2 diabetes mellitus
|
Study design
Specification of the type of study design(s)
- Case-control
- Cohort
- Cross-sectional
- Descriptive or case series
- Clinical trial
- Population screening
|
- Case-control Study
|
Case definition
For study designs 1, 4, and 5, define the following if available:
- Disease case definition
- Exclusion criteria
- Gender
- Race/ethnicity
- Age
- Time period
- Geographic location
- Number of participants
|
- Disease case definition: WHO criteria for diabetes: plasma glucose concentration = 126 mg/dl after fasting or = 200 mg/dl 2 hours after 75 g oral glucose load in oral glucose tolerance test (OGTT). Type 2 DM defined as not requiring insulin within 12 months of diagnosis.
- Exclusion criteria: Type 2 DM only (not Type 1)
- Gender: Both
- Race/ethnicity: Caucasian
- Age: 66.8 mean (10.9 SD)
- Time period: 2001-2002
- Geographic location: Recruited from University College Diabetes and Cardiovascular Disease Study whose subjects are from the diabetes clinic at University College London Hospitals NHS Trust.
- Number of participants: 574/605 (95 % of total eligible)
|
Control definition
For study design 1, define the following if available:
- Control selection criteria
- Matching variables
- Exclusion criteria
- Gender
- Race/ethnicity
- Age
- Time period
- Geographic location
- Number of participants
|
- Control selection criteria: Recruited from Northwick Park Heart Study II: unrelated healthy males recruited from 9 UK general medical practices throughout UK and prospectively followed for development of CHD since 1989.
- Matching variables: Race
- Exclusion criteria: No diabetes. Diabetes status obtained by direct questioning of symptoms and previous diagnosis in order to exclude those with diabetes. (No biochemical diagnosis.)
- Gender: Male
- Race/ethnicity: Caucasian
- Age: 56.0 mean (3.5 SD)
- Time period: 1989-?
- Geographic location: 9 UK general medical practices throughout UK Number of participants: 2413/3012 (80 % of total eligible)
|
Assessment of environment factors
For studies that include gene-environment interactions, define the following, if available:
- Environmental factor
- Exposure assessment
- Exposure definition
- Number of participants with exposure data (% of total eligible)
|
- Environmental factor: N/A
- Exposure assessment:
- Exposure definition:
- Number of participants with exposure data: N (% of total eligible)
|
Genotyping
Specify the following:
- Gene
- DNA source
- Methodology
- Number of participants genotyped (% of total eligible)
|
- Gene: Angiotensin-1 converting enzyme (ACE)
- DNA source: Genomic leukocyte DNA from blood
- Genotyping method: PCR using the three primer method (two oligonucleotide primers outside the insertion sequence and one primer inside the sequence) with amplification products of 65 bp (I allele) and 84 bp (D allele). (Inner primer specific to insertion so smaller product preferably amplified when insertion present vs. only outer primers used when deletion present.) PCR product sizes determined using microtiter array diagonal gel electrophoresis (MADGE).
- Number of participants genotyped: 2987/3617 (83% of total eligible)
|
Analysis
Comment on the analysis carried out by the author(s), e.g. matching, modeling or statistical tests used. Were the analyses appropriate?
|
Cases included both sexes but controls were male only. Cases older on average than controls. Some non-diabetic controls may develop Type 2 DM by the time average age reaches that of cases since incidence rates increase with age. |
Results
Describe the major results under each of the following HuGE categories. Include tables when data are provided:
- Prevalence of gene variant
- Gene-disease association
- Gene-environment interaction
- Gene-gene interaction
- Genetic test evaluation/monitoring
|
Genotype |
Cases |
Controls |
OR |
95 % C.I. |
II |
62 (17.4%) |
568 (23.5%) |
1.00 |
- |
ID |
188 (52.8%) |
1183 (49.0%) |
1.46 |
1.07 - 1.97 |
DD |
106 (30.0%) |
662 (27.4%) |
1.47 |
1.05 - 2.05 |
p |
|
|
0.03 |
|
II |
62 (17.4%) |
568 (23.5%) |
1.0 |
- |
ID, DD |
294 (82.6%) |
1845 (76.5%) |
1.46 |
1.10-2.32 |
p |
|
|
0.01 |
|
OR for family history of diabetes in DD compared to II = 1.52 [0.89-2.60], p=0.03
Table 3. Odds ratio for a family history of diabetes mellitus by ACE (I/D) genotype in UDACS
| |
Family history/total |
OR (95% CI) |
II |
28/98 (28.6%) |
1.00 |
ID |
71/267 (26.6%) |
0.91 (0.54–1.52) |
DD |
67/177 (37.9%) |
1.52 (0.89–2.60) |
P=0.03 |
|
|
There was a significant association between genotype and family history of diabetes (p = 0.03). DD subjects had a significantly higher OR for a family history (when DD were compared to grouped II/ID, the OR was 1.63, p = 0.01).
Also, no association seen between ACE I/D and CHD or proteinuria (used as a measure of nephropathy).
For plasma CRP, only significant difference seen for those not on ACE inhibitors with DD genotype (compared to ID/II genotypes) = 1.90 ± 1.66 mg/L vs. 1.56 ± 1.29 mg/L, p = 0.03 adjusted.
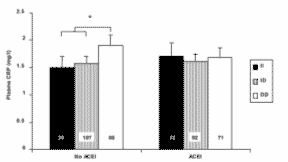
|
Conclusion
State the author's overall conclusions from the study |
Authors state that "this study clearly shows an association" between the ACE D allele and Type 2 DM. |
Comments
Provide additional insight, including methodologic issues and/or concerns about the study |
**Caveats to the following calculations:
- Substitution of OR for RR is less accurate in this case since both the D allele and type 2 diabetes are not rare.
- Sample is not population-based.
- The causal relationship has not been established between the presence of the D allele and type 2 diabetes.
Attributable fraction (using OR for RR) for D carriers (Men only):
(RR-1)/RR = (1.55-1)/1.55 = 35.4 %
Attributable fraction in population (Men only)
fc = 294/356 = 0.826
fc(RR-1)/RR = (0.826)(1.55-1)/1.55 = 29.3 %
Population screening is not recommended as most people identified through screening would not develop Type 2 DM (at least not due to their ACE genotype).
Other factors such as health behaviors (BMI, diet, physical activity) and family history offer much more utility in terms of predicting onset of Type 2 DM. While treatment with ACE inhibitors may potentially be helpful in the future for D carriers with Type 2 DM, such benefits may be limited to a subset of patients and must be investigated further.
|


