Science Update: Food Safety Help for Processors
A new Internet resource could be invaluable to food processors—especially, to smaller meat and poultry processing companies. The information it provides can answer food safety questions and help food processors make science-based decisions to produce safe and wholesome products for consumers. Called the Predictive Microbiology Information Portal, or PMIP, it offers information on research, regulations, and resources related to Listeria monocytogenes in ready-to-eat foods. Soon it will be expanded to include other pathogen and food combinations. To help ensure the safety of food processing methods, a searchable database allows users to find information that can be used to develop plans for Hazard Analysis and Critical Control Point inspection.
Developed in collaboration with Rutgers University and Decisionalysis Risk Consultants, Inc., of Ottawa, Ontario, the portal includes a tutorial with instructions on using and interpreting predictive models. It links to the ARS Pathogen Modeling Program and ComBase, which provide diverse resources associated with databases, regulatory requirements, and food safety principles. The PMIP Web address is www.ars.usda.gov/naa/errc/mfsru/portal.
Vijay K. Juneja, USDA-ARS Microbial Food Safety Research Unit, Wyndmoor, Pennsylvania; phone (215) 233-6500.
_____________________________________
USDA Web Portal Offers Big Food Safety Benefits for Small Food Processors
by Laura McGinnis
September 19, 2007

Electron micrograph of a Listeria monocytogenes bacterium in tissue. The new web portal will help food processors protect food from Listeria and Similar pathogens. Image courtesy Public Health Image Library of the Centers for Disease Control and Prevention.
WASHINGTON, September 19, 2007 - The U.S. Department of Agriculture (USDA) has unveiled a new Internet resource to help small companies answer food safety questions and help food processors make science-based food production decisions. The Internet portal, available at http://www.ars.usda.gov/naa/errc/mfsru/portal, is one of the most comprehensive decision support tools available.
Read more about this topic.
__________________________________
Agricul tural Research Service (ARS) Scientists Honored for Food Safety Research

On August 16, the International Association for Food Protection (IAFP) is honoring scientists in the Microbial Food Safety Research Unit (MFSRU), Eastern Regional Research Center (ERRC), Wyndmoor, PA, with the 2006 Food Product Association Food Safety Award for their research over the past 2 decades to enhance the safety and security of our food. IAFP is an internationally renowned organization comprising over 3,000 members from 50 countries. John P. Cherry, Center Director, ERRC, and John B. Luchansky, Research Leader, MFSRU, will accept the award on behalf of the ARS scientists at the IAFP 2006 53rd Annual Meeting in Calgary, Alberto, Canada.
Unraveling the Listeria Genome
If knowledge is power, Agricultural Research Service scientists are gaining the upper hand on Listeria monocytogenes.
Researchers at the ARS, Eastern Regional Research Center (ERRC) in Wyndmoor, PA and the Institute for Genomic Research in Rockville, Maryland, sequenced four strains of Listeria related to food borne listeriosis outbreaks - an important step toward developing a management strategy for this deadly bacterium.
Read more about the research in the October 2006 issue of Agricultural Research Magazine.
Current Research Projects at MFS
Adoption of USDA/ARS Method to Extract Noroviruses from Oysters
For additional information contact:
Gary P. Richards
Gary.Richards@ars.usda.gov
tel: 302-857-6419
Fax: 302-857-6451
In February, 2006, Health Canada published a procedure to extract and assay noroviruses in oysters in heir Compendium of Analytical Methods (http://www.hc-sc.gc.ca/fn-an/res-rech/analy-meth/microbio/volume5/index_e.html). The virus extraction method is based on a procedure published in 2001 by Drs. David H. Kingsley and Gary P. Richards (Kingsley, D.H., and G.P. Richards. 2001. Rapid and efficient method for reverse transcription-PCR detection of hepatitis A and Norwalk-like viruses in shellfish. Applied and Environmental Microbiology Vol. 67, pp. 4152-4157). Seven laboratories in Canada and three in the United States participated in testing the method including laboratories at Health Canada, the Canadian Food Inspection Agency, Agriculture and Agri-Food Canada, the British Columbia (BC) Centre for Disease Control, the Centre québécois d’inspection des aliments et de santé animale, the University of California at Davis, the U.S. Food and Drug Administration, and the Alaska Environmental Health Laboratory. The Compendium of Analytical Methods is a reference of methods used by Health Canada, Agriculture and Agri-Food Canada, and the Canadian Food Inspection Agency. Noroviruses, the primary cause of food-related illness in North America, have been associated with outbreaks in schools, hospitals, and nursing homes, and on cruise ships. They are frequently associated with the consumption of contaminated water or food. Oysters and clams have been associated with numerous large outbreaks of norovirus illness. Symptoms of this "stomach virus" include nausea and potentially severe diarrhea and/or vomiting developing usually within a day after consuming food or drink that is contaminated with fecal material. Canadian scientists first contacted ARS about a suspected outbreak of oyster-associated norovirus illness in British Columbia, which led to an evaluation of the ARS method. At the request of the U.S. FDA, Drs. Kingsley and Richards, and Gloria Meade used the method to successfully detect hepatitis A virus and norovirus in imported clams implicated in an outbreak of norovirus illness in New York State (Kingsley, D.H., G.K. Meade, and G.P. Richards. 2002. Detection of both hepatitis A and Norwalk-like virus in imported clams associated with food-borne illness. Applied and Environmental Microbiology Vol. 68, pp. 3914-3918).
Development of a New Vibrio Detection Assay: The Colony Overlay Procedure for Peptidases
In the United States, Vibrio vulnificus and Vibrio parahaemolyticus are the major shellfish-borne bacterial pathogens and are naturally present in shellfish-harvesting waters and in oysters, clams, and mussels. Vibrio vulnificus is particularly deadly, with mortality rates approaching 60% among those who become ill. Diabetics and those with liver disease who eat raw or undercooked shellfish are at the highest risk of V. vulnificus infection and death. Vibrio parahaemolyticus outbreaks have been responsible for massive closures of shellfish harvesting areas along the Atlantic, Gulf, and Pacific coasts of the U.S. A third species of Vibrio, known as V. cholerae, causes highly infectious cholera in developing countries. Dr. Gary Richards, Michael Watson, and Dr. Salina Parveen developed and published an enzyme assay known as the colony overlay procedure for peptidases or COPP assay for short (Richards, G.P., M.A. Watson, and S. Parveen. 2005. Development of a simple and rapid fluorogenic procedure for identification of Vibrionaceae family members. Applied and Environmental Microbiology Vol. 71, pp. 3524-3527). The COPP assay is a simple plating procedure that requires the overlay of bacterial colonies with a cellulose acetate membrane that contains a fluorescent substrate. The assay detects and quantifies Vibrionaceae family members in seawater and shellfish based on the presence of an enzyme, which is only found in Vibrionaceae family members. This procedure is relatively simple, rapid, and inexpensive. Other Vibrio testing methods often require enrichment procedures, multiple tube (most-probable-number) assays, and complex media and are usually designed to detect specific pathogenic species, not total Vibrionaceae.
Over the past two years, Dr. Richards has collaborated with Dr. David Bushek and Iris Burt from the Haskin Shellfish Research Laboratory, Rutgers University, Port Norris, NJ, in evaluating the use of the COPP assay for total Vibrionaceae detection in seawater and shellfish obtained during monthly surveys in the Delaware Bay. Counts exceeding a million Vibrionaceae per gram of shellfish are commonly encountered during the summer months. Studies are underway to biochemically identify the Vibrio species isolated. High levels of total Vibrionaceae may serve as an indicator for the possible presence of pathogenic Vibrio species, much like high fecal coli form levels are used by regulators to restrict shellfish harvesting. The COPP assay provides, for the first time, an opportunity to determine relationships between total Vibrionaceae counts and the incidence of Vibrio outbreaks in affected areas. The assay may also be practical for monitoring spikes in total Vibrionaceae counts in aquaculture settings where spikes may signify an active disease outbreak or a period when the animals are stressed, perhaps from poor water quality or other system failures.
Recently, Dr. Richards and his collaborator Dr. Alberto Nuñez from the ARS Eastern Regional Research Center in Wyndmoor, PA, determined that the enzyme that produces the fluorescence on the COPP assay (phosphoglucose isomerase with a lysyl aminopeptidase activity) may also enhance virulence of at least some of the Vibrionaceae through the proteolytic processing of human derived peptides (Richards, G.P. and A. Nuñez. 2006. Specificity of a Vibrio vulnificus aminopeptidase toward kinins and other peptidyl substrates. Journal of Bacteriology Vol. 188, pp. 2056-2062). The products produced by the proteolytic processing of these peptides are known to enhance inflammatory reactions, vasodilation, and vascular permeability in infected individuals. This finding may help explain the rapid progression and high mortality rate for V. vulnificus infections. For more information contact:
Gary P. Richards, Ph.D
Lead Scientist
USDA, ARS
Delaware State University
James W.W. Baker Center
Dover, DE 19901
Phone: 302-857-6419
Fax: 302-857-6451
Gary.Richards@ars.usda.gov
David Kingsley, Ph.D.
Microbiologist
Delaware State University
James W.W. Baker Center
Dover, DE 19901
Phone: 302-856-6406
Fax: 302-857-6451
David.Kingsley@ars.usda.gov
Support personnel:
Michael Watson
Delaware State University
James W. Baker Center
Dover, DE 19901
Phone: 302-857-7881
Fax: 302-857-6451
Michael.Watson@ars.usda.gov
Gloria Meade
Delaware State University
James W. Baker Center
Dover, DE 19901
Gloria.Meade@ars.usda.gov
(back)
Molecular Characterization of Pathogens and Their Responses to Environmental Factors
For additional information contact:
Pina M. Fratamico
pina.fratamico@ars.usda.gov
tel: 215-233-6525
Fax: 215-233-6581
Program Background
Food and agriculture systems expose pathogenic bacteria to heat, cold, salt, acid, oxidative, and other stresses that provoke bacterial responses designed to cope with the particular stress situation. Bacteria possess mechanisms to adapt to sublethal stresses, rendering them resistant to lethal levels of the same stress and/or resulting in the capacity to withstand other types of stresses. This is important because stress-adapted bacteria may be more difficult to inactivate with techniques commonly used by the food industry. Stresses used in food systems to control bacterial growth may, in fact, increase the disease-causing (virulence) potential of pathogens that survive the particular treatment. In addition, surface structures such as “curli”, which are involved in biofilm formation, are controlled by environmental signals. Using genomics (study of genes) and proteomics (study of proteins) tools, we are investigating the regulatory networks that allow pathogens to survive despite the growth barriers placed in foods and during food processing.
Quorum sensing is a phenomenon through which small signaling molecules termed autoinducers are employed for bacterial cell-to‑cell communication in response to population density and environmental stresses. Studies have shown that quorum sensing is widespread and involves complex interconnected regulatory networks that serve to fine tune the expression of diverse behaviors, including sporulation, virulence, and biofilm formation. Our research focuses on elucidating how conditions in food environments initiate quorum sensing responses, which then modulate pathogen behavior in favor of survival.
In addition, genomics/proteomics approaches are being used for molecular characterization of pathogens to understand their virulence potential and how they develop resistance to food-related stresses, and they are also being used to identify molecular markers for detection, identification, and typing of pathogens. These research approaches provide fundamental information for the development of intervention strategies for the control of pathogens in foods.
Research Objectives
(1) Investigate the mechanisms for pathogen adaptation in foods to determine what conditions affect survival, resistance, and pathogenicity.
(2) Investigate bacterial cell-to-cell signaling mechanisms (quorum sensing) and their role in growth, survival, biofilm formation, and pathogenicity in foods.
(3) Develop and utilize genetic-based methods for identification, typing, and characterization of food-borne pathogens.
Pathogens of interest are enterohemorrhagic Escherichia coli O157:H7 and other pathogenic E. coli, Campylobacter species, and Salmonella species.
Campylobacter jejuni is a microaerobic organism that prefers lower levels of oxygen than that found in air. We are interested in understanding how C. jejuni regulates its ability to tolerate stresses and how the pathogen is able to survive and grow when exposed to air or other oxidative stresses that might be found during food processing and storage. We are also interested in population density effects (quorum sensing) on expression of genes involved in virulence, growth, and survival. In addition, Campylobacter is able to survive for long periods of time at refrigeration temperature (ca. 4°C), and we are examining the genes/proteins involved in regulating the tolerance of Campylobacter to cold temperatures. Various approaches, including the use of microarrays and two-dimensional gel electrophoresis are used to study global gene/protein expression patterns under the above-mentioned conditions. Real-time PCR, microbial genetics, and other molecular techniques are used to study specific gene targets that are implicated by the global expression analyses or that are inferred from data from other bacterial pathogens. Our ability to readily identify genes and proteins that are responsible for the ability of pathogens to survive environmental insults will allow us to ultimately circumvent the survival mechanism(s) of food-borne pathogens and ultimately enhance food safety.
We have conducted a proteomic survey of Escherichia coli O157:H7 strain EDL933 using two-dimensional gel electrophoresis and a column-based high pressure liquid chromatography protein separation technique, comparing the parent strain to its mutant variant. The mutant has a single base pair mutation in the promoter region of the csgD gene and produces curli structures, is invasive, forms thicker biofilms than the parent strain, and is more infective in animal models. One of the goals of this work is to identify candidate proteins that play a role in the ability of E. coli O157:H7 to form biofilms. Both techniques produced sets of proteins, which were differentially expressed in the mutant as compared to the parent strain. One of these proteins was found to be highly expressed in the parent strain but was absent in the mutant. Work is underway to determine the exact role of this and other identified proteins in the survival of E. coli O157:H7 and in biofilm formation. The proteomic analysis has also identified potential oxidative stress genes, which may contribute to increased stress tolerance of pathogenic E. coli residing in biofilms. We are using deletion analysis to determine the contribution of various E. coli genes to its resistance to oxidative stresses.
Genetic methods with the ability to precisely discriminate among different species of Campylobacter were developed. These methods will be essential for effective monitoring and surveillance to determine the prevalence of these organisms in food, in animals, and in the environment and for defining the epidemiology of human infections. In addition, a multiplex real-time polymerase chain reaction (PCR) method was developed for detection of E. coli O157:H7 in food and other types of samples. PCR assays for detection, identification, and typing of other pathogens have also been developed. Finally strains of E. coli and Salmonella isolated from food and animals were characterized by determining the presence of virulence genes, their antibiotic resistance profiles, and the ability of the strains isolated from animals and food to tolerate stresses to understand the potential of these organisms to cause human illness.
Impact
The research on bacterial stress responses provides information on the effects of food environments and food processing and storage conditions on growth and survival of pathogenic bacteria that is necessary for the development of strategies to decrease viability of pathogens in foods. The research on quorum sensing will assist in understanding how biofilms are formed. It also provides information on the types and levels of cell-to-cell signaling molecules produced by food-borne pathogens under different conditions and on bacterial processes that are modulated by these signals. Control strategies that modulate the production or that inactivate signaling molecules may offer a means to control the growth of undesirable bacteria in food. Finally, new approaches and tools to type and characterize emerging pathogens, including new serogroups of E. coli associated with diarrheal illness, non-jejuni/coli species of Campylobacter, and antibiotic resistant organisms are essential for identification and control of new threats, including agents that may be employed for bioterrorism.
 |
Biofilm formation of E. coli O157:H7
strains 43895OR (OR; curli-producing) and 43895OW
(OW; non-curli-producing) on glass, Teflon, and
stainless steel surfaces. |
 |
Topographical images of colonies of E. coli O157:H7 strains (A) 43895OW (curli non-producing) and (B) 43895OR (curli producing) grown on agar for 48 h at 28oC. (C) Negatively stained transmission electron microscopy image of strain 43895OR showing curli (double arrow). (D) Immunoblot of proteins of strains 43895OR (Lane 1) and 43895OW (Lane 2) reacted with anti-curli antibody. Only strain 43895OR expresses a protein that reacts with the antibody (double arrow). |
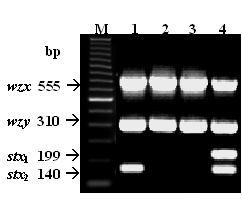 |
Agarose gel showing results of a multiplex polymerase chain reaction (PCR) assay targeting the E. coli O145 wzx and wzy genes (found in the E. coli O145 O antigen gene cluster) and the Shiga toxin genes. All 4 strains were positive for wzx and wzy, in addition to either the Shiga toxin 2 (lane 1) or Shiga toxin 1 and 2 genes (lane 4). M, molecular weight standards. |
Collaborations
The Pennsylvania State University
The University of Naples
Istituto Superiore di Sanitá (Rome)
National Food Research Institute (Japan)
Centro Universitario, Querétaro, México
EMBRAPA, Brazil
INIFAP, Mexico
CIAD, Mexico
Purdue University
Ursinus College
Principal Investigators
Pina M. Fratamico, Ph.D.
Microbiologist/Lead Scientist
tel. 215-233-6525
fax. 215-233-6581
pina.fratamico@ars.usda.gov
Chin-Yi Chen, Ph.D.
Molecular Biologist
tel. 215-233-6526
fax. 215-233-6581
chin-yi.chen@ars.usda.gov
Gaylen Uhlich, Ph.D.
Molecular Biologist
tel. 215-233-6740
fax. 215-233-6581
gaylen.uhlich@ars.usda.gov
Nereus “Jack” Gunther, Ph.D.
Molecular Biologist
tel. 215-233-6503
fax. 215-233-6581
jack.gunther@ars.usda.gov
Xianghe Yan, Ph.D.
Computational Biologist
tel. 215-233-6732
fax. 215-233-6581
xianghe.yan@ars.usda.gov
Project Personnel
Lori K. Bagi
Microbiologist
tel. 215-233-6624
fax. 215-233-6581
lori.bagi@ars.usda.gov
Bryan Cottrell
Biological Science Laboratory Technician
tel. 215-836-3751
fax. 215-233-6581
bryan.cottrell@ars.usda.gov
Terence P. Strobaugh
Biological Science Laboratory Technician
tel. 215-233-6455
fax. 215-233-6581
terence.strobaugh@ars.usda.gov
Jonnee Almond
Biological Science Laboratory Technician
tel. 215-233-6410
fax. 215-233-6581
jonnee.almond@ars.usda.gov
(back)
Effect of Interventions and Processes on Persistence of Pathogens on Foods
For additional information contact:
John Luchansky
John.Luchansky@ars.usda.gov
tel: 215-233-6620
Program Background
The focus of this research is to identify where pathogens enter the food supply, how they persist, and how to eliminate or control them. The targeted pathogens are Listeria monocytogenes, Salmonella, Escherichia coli O157:H7, and Bacillus anthracis while targeted foods are ready-to-eat meat, poultry products, fluid milk, and Hispanic-style cheese. We are working to identify the sources of these pathogens in foods and in food processing environments, and to determine the genes and proteins that are responsible for pathogen survival and persistence. DNA fingerprinting is used to identify pathogens from sources between the farm and retail distribution to determine where pathogens persist. Processes and interventions, such as fermentation, microfiltration, high pressure processing, food grade chemicals, and heating, alone or in combination, are evaluated for their ability to inhibit or remove undesirable bacteria and to better manage pathogen presence and/or survival during manufacture and storage. The research results are needed to assist manufacturers in meeting current regulatory guidelines and to assist regulators in science-based policy decisions. Using the tools of genomics and proteomics, we are working to identify the genes and proteins necessary for pathogens to survive in food-related environments and that are responsible for human food-borne illnesses. These studies help us understand significant food borne pathogens and bioterrorism threat agents, and will lead to better methods for controlling these organisms in foods prior to human contact and consumption. Ultimately this enhances the safety and security of our food supply.
Research Objectives
The overall objective is to reduce the occurrence, risk, and severity of human illness resulting from the consumption of foods contaminated with pathogenic microorganisms. The project focuses on the following three main objectives aimed at increasing our understanding of pathogen persistence in foods and in turn developing and evaluating effective interventions to enhance the safety and security of our Nation’s food supply:
• Determine the ecology of pathogens in various foods with specific focus on foods considered high risk by food industry regulators like the Food Safety Inspection Service and the Food and Drug Administration. Example foods include ready-to-eat foods, or foods with a short shelf life.
• Develop and validate intervention strategies to be used alone or in combination with other processes for pathogen control.
• Using genomic and proteomic approaches, determine the pathogens’ physiological responses to various intervention strategies, food processes, and to the inherent food production macro and microenvironments.
Impact
The research on molecular characterization of pathogens and threat agents using techniques such as microarrays and analysis of genetic mutants provides important information regarding the underlying genetic basis for addressing why certain strains or serotypes are more prominent in food borne outbreaks and why certain strains or serotypes survive better within different foods and in the processing environment.
DNA fingerprinting methods are applied throughout the farm-to-table chain to determine entry points, incidence and persistence of pathogens on higher-risk foods. This research will also update information on the types and subtypes of pathogens on foods and result in use of relevant strains in intervention studies.
Finally, this research evaluates the efficacy of processing methods and interventions on pathogen persistence. This aspect of the research can have immediate impact by identifying new approaches for reducing pathogen levels on higher-risk foods. Some examples are:
• Inclusion of potassium lactate as an ingredient in packages of frankfurters is sufficient to prevent outgrowth of the pathogen Listeria monocytogenes during 90 days of refrigerated storage. This finding had immediate impact since manufacturers using potassium lactate as an ingredient could satisfy “Alternative 2” of the latest USDA FSIS policy by preventing outgrowth of this pathogen and, as such, are not subjected as frequently to regulatory sampling.
• Pasteurization of milk does not inactivate spores of the threat agent Bacillus anthracis. In collaboration with the Dairy Processing and Products Research Unit, new technologies are being developed that remove spores and pathogenic bacteria from milk while maintaining the quality and nutritional profile of milk. The technologies are validated at a scale that is relevant to commercial operation and are designed to fit into existing fluid milk processing lines to minimize capital and operating costs. Prefatory experiments demonstrated removal of 99.9999% of B. anthracis spores from a skim milk stream. This research will have immediate impact as a new approach for improving the security of the milk supply.
Principal Investigators
John B. Luchansky, Ph.D
Lead Scientist, MFSRU
tel. 215-233-6620
fax. 215-233-6581
John.Luchansky@ars.usda.gov
Peggy Tomasula, D.Sc
Research Leader, DPPRU
tel. 215-233-6703
fax. 215-233-6795
Peggy.Tomasula@ars.usda.gov
Yanhong Liu, Ph.D.
Research Microbiologist
tel. 215-233-6587
fax. 215-233-6581
Yanhong.Liu@ars.usda.gov
Sudarsan, Mukhopadhyay, Ph.D
Research Chemical Engineer
Tel. 215-233-6600
Fax. 215-233-6581
Sudarsan.Mukhopadhyay@ars.usda.gov
Support Scientist
Jeffrey Call
Supervisory Microbiologist
Tel. 215-233-6789
Fax. 215-233-6581
Jeffrey.Call@ars.usda.gov
Postdoctoral Fellows
Anna PortoFett, Ph. D
Research Food Toxicologist
Tel. 215-836-3762
Fax. 215-233-6581
Anna.PortoFett@ars.usda.gov
Amy Ream, M.S.
Microbiologist
Tel. 215-233-6760
Fax. 215-233-6581
Amy.Ream@ars.usda.gov
(back)
Microbial Modeling & Bioinformatics for Food Safety and Security
For additional information contact:
Vijay Juneja
vijay.juneja@ars.usda.gov
tel: (215) 233-6500
Program Background
Understanding the risks of bacterial foodborne illness depends on knowledge of specific environmental conditions that affect pathogen behavior (i.e., growth, survival and decline). This information can be translated into mathematical models that allow food safety managers to estimate how specific pathogens will react to unique environmental conditions.
Such models are used by food industries to develop new product formulations that minimize pathogen growth, and to design and implement Hazard Analysis & Critical Control Points (HACCP) food safety systems. Predictive models also provide risk analysts with efficient tools to define intervention strategies and policies that reduce the risk of foodborne disease.
The ARS Pathogen Modeling Program (PMP; http://www.arserrc.gov/mfs/pathogen.htm) is a software package of microbial models and a research product of the Microbial Food Safety Research Unit (MFS) that is meeting the needs of ARS customers in government, industry and academia. The PMP contains models that allow users to predict food formulation, processing and handling conditions that control the growth, survival and death of various bacterial foodborne pathogens.

ComBase (www.combase.cc), a collaborative project of the USDA Agricultural Research Service, Eastern regional research Center, UK Institute of Food Research and the Australian Food Safety Center of Excellence. It contains over 35,000 datasets that describe the growth, survival and inactivation of bacteria under diverse environments relevant to food processing operations. This tool brings vast amounts of data to food safety managers and risk assessors for estimating the fate of bacteria in diverse types of foods.
Research Objectives
The Microbial Modeling & Bioinformatics for Food Safety & Security research team is addressing the following high priority research topics:
• Developing and validating new predictive models for estimating the responses of microbial pathogens in select food matrices, as a function of food formulation, competitive flora, thermal inactivation, and process unit operations.
• Producing new and objective measures for assessing model performance and robustness.
• Determining strategies that can be used to reduce the uncertainty of predicting bacterial lag times.
Impact
This research has resulted in the following accomplishments:
• Models that predict the growth of Clostridium perfringens and Clostridium botulinum in ready-to-eat meats, which provide the scientific basis for regulatory performance standards/compliance guidelines.
• Growth models for Escherichia coli O157:H7 in raw ground beef that are assisting food companies in meeting regulatory standards.
• Models that help food safety managers control the growth of Listeria monocytogenes in delicatessen salads (seafood, ham, potato) under refrigeration and temperature-abuse conditions.
These research products are used extensively by the food industry and the USDA Food Safety & Inspection Service to design, implement and evaluate HACCP plans. Each year, the PMP is downloaded from the MFS website by more than 5,000 users in numerous countries. These predictive models also fill important risk assessment knowledge gaps, thus producing more accurate estimations of foodborne hazards. Current initiatives also include enhancing the PMP and ComBase software so that they better assist small and very small food companies in meeting food regulations and performance standards.
Principal Investigators
Vijay Juneja, Ph.D.
Lead Scientist/ Supervisory Microbiologist
Tel. 215-233-6500
Fax. 215-233-6697
vijay.juneja@ars.usda.gov
Saumya Bhaduri, Ph.D.
Microbiologist (Molecular Biology)
Tel. 215-233-6521
Fax. 215-233-6581
saumya.bhaduri@ars.usda.gov
Andy Hwang, Ph.D.
Research Food Technologist
Tel. 215-233-6416
Fax. 215-233-6581
andy.hwang@ars.usda.gov
Shiowshuh Sheen, Ph.D.
Research Food Technologist
Tel. 215-836-3774
Fax. 215-233-6581
shiowshuh.sheen@ars.usda.gov
Thomas P. Oscar, Ph.D.
Research Food Technologist
Tel. 410-651-6062
Fax. 410-651-8498
Thomas.Oscar@ars.usda.gov
toscar@umes.edu
Jacquelyn Ludwig
Biological Science Laboratory Technician
Tel. 410-651-6372/7235
Fax. 410-651-6568
Jacquelyn.Ludwig@ars.usda.gov
Kenyetta Chaney, B.S.
Biological Science Laboratory
Technician
Tel. 215-836-3749
Fax. 215-233-6581
kenyetta.chaney@ars.usda.gov
Angie Martinez, B.S.
Biologist
Tel. 215-233-6425
Fax. 215-233-6581
angie.martinez@ars.usda.gov
Stacy Raleigh, B.S.
Biological Science Laboratory Technician
Tel. 215-233-6437
Fax. 215-233-6581
Stacy.Raleigh@ars.usda.gov
(back)
|