 |
|
DNA Repair & Mitochondrial Damage Group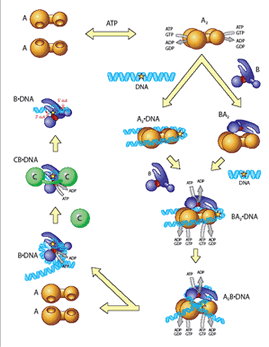
Figure 1: A diagram of the nucleotide excision repair pathway.
Faulty DNA repair can promote mutations, aging, cancer and cell death. The process by which protein components of repair detect damaged or modified bases within DNA is an important but poorly understood type of protein-DNA interaction. One of the most remarkable aspects of nucleotide excision repair (NER) is that it can remove a wide range of DNA lesions that differ in chemistry and structure. During bacterial NER UvrA, UvrB (http://pdbbeta.rcsb.org/pdb/explore.do?structureId=1t5l) To overcome this problem, UvrA, UvrB and UvrC from the thermophilic bacteria Bacillus caldotenax and Thermotoga maritima were recently cloned and overexpressed. The proteins maintain optimal activity at 65ºC and are amenable to both structural and biophysical studies. The group is collaborating with Bob London’s NMR group at NIEHS for an analysis of the dynamics of UvrB upon ligand binding using NMR techniques. The group is also collaborating with Caroline Kisker, Ph.D. and her group, in Wurtzburg, Germany, on solving protein and protein-DNA structures by X-ray crystallography. These complexes are being visualized on DNA using atomic force microscopy, performed by Dorothy Erie, Ph.D., at UNC, and single-molecule techniques using quantum dot labeling, performed by David Warshaw, Ph.D. at the University of Vermont. These tools, combined with site-directed mutagenesis and biochemical analyses, allow for structure-function studies of the UvrA, UvrB and UvrC proteins, and form a basis for understanding the fundamental molecular processes of NER. The long-term goal is to have a complete understanding of how structural perturbations induced by specific DNA lesions are detected and removed by the NER machinery at the atomic level. |
|

