|
|
||||
| NCBI Map Viewer is a Web resource that anyone can use to view and search an organism's complete genome. Users can also view maps of individual chromosomes and zoom into specific regions within chromosomes to explore the genome at the sequence level. Map Viewer provides access to several different types of maps for different organisms. Many of these maps are meaningful only to scientific researchers. A discussion of all the different types of maps and genomic data is beyond the scope of this tutorial, which will focus only on how to locate a specific gene on a chromosome map. For demonstration purposes, we will search for the chromosomal location of the hereditary hemochromatosis gene, HFE. The same process can be carried out for any gene. Before you begin to explore Map Viewer or other molecular databases to retrieve information about a particular gene, you should know the official symbol for your gene and the number of the chromosome on which it resides. One resource for finding this information about a gene is Online Mendelian Inheritance in Man (OMIM). For tips on searching OMIM see our tutorial. To begin, go to the
NCBI Map Viewer home page 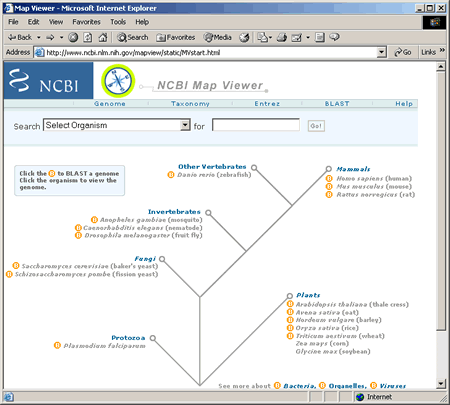
To access the genome view for humans, select Homo sapiens (humans) from the Search drop down box and click Go!, or simply click on Homo sapiens (humans) under Mammals in the tree structure on the home page. The Homo sapiens genome view is shown in the following screenshot. 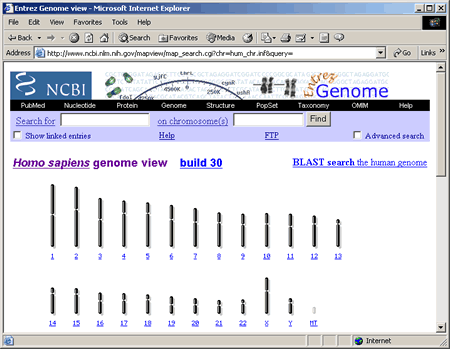
After using OMIM,
we determined that the gene for the most common form of hereditary hemochromatosis
resides on chromosome 6 and that the symbol for this gene is HFE. In the
Search for
box at the top of the page, enter the gene symbol HFE and, in the
on chromosome(s)
box, enter 6 as demonstrated below. The Search
for and
on chromosome(s)
links take you to the section of the Entrez
Map Viewer Help Document that describes these search features. Click
This search returned one hit on chromosome 6 (as indicated by the red number 1 listed below the chromosome number). The genome view now displays one red mark on chromosome 6 marking the approximate location of the HFE gene.
Below the genome view is a table describing the match to your query.
The maps and map element, which also serve as links to maps of regions on chromosome 6, are described below. Due to differences in map and display settings, the view of the chromosome associated with each link will vary.
To access a map of chromosome 6, click on the the Genes_cyto link. This opens a view of Chromosome 6 that should resemble the following screenshot. 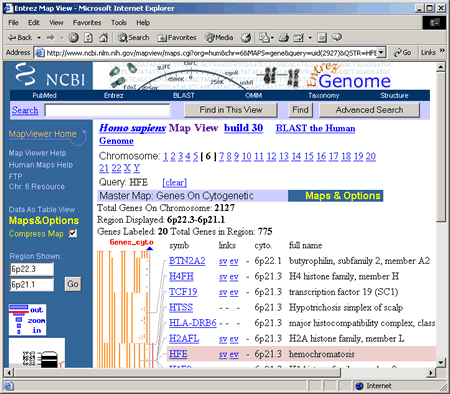 Take some time to explore the different features of this Chromosome View. The particular type of map displayed in this view is the Genes on Cytogenetic map. Note that you can directly link to maps of other chromosomes by simply clicking on a chromosome number. Each chromosome view gives the total number of genes that have been mapped to that chromosome and identifies the specific region of the chromosome displayed in the view. For this view, the region between 6p22.3 and 6p21.1 is shown. The small chromosome image in the blue column on the left shows where this region is located on the chromosome. Both 6p22.3 and 6p21.1 are cytogenetic map locations. For the location 6p21.1, 6 is the chromosome number, p indicates the short arm of the chromosome, and 21.1 is the number assigned to a particular band on a chromosome. When chromosomes are stained in the lab, light and dark bands appear, and each band is numbered. The higher the number, the farther away the band is from the centromere. More information about display options and map views is provided in the next section of this tutorial. Users
can zoom in on a particular region of a chromosome, select a particular
map type to serve as the Master Map, and adjust other display options
by clicking on the 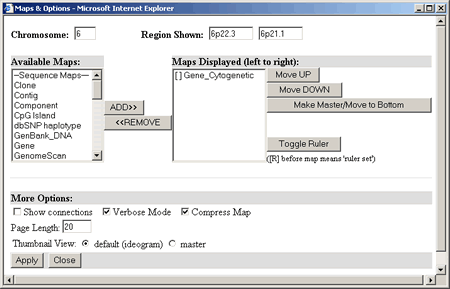
Select the following options to modify the map display:
Select 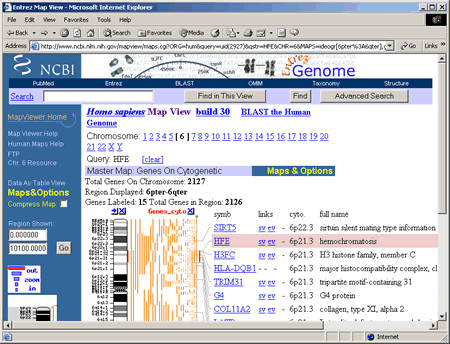
The red marks on the ideogram and Genes_cyto maps show where the HFE gene can be found. The Genes_cyto map, which displays the cytogenetic locations of genes listed in NCBI's LocusLink, is our master map. The HFE gene is highlighted in pink because it matched our query term, HFE. The HFE map element in the Genes_cyto map includes the following links to additional information:
To learn more about what you can do with Map Viewer, experiment with different features on your own or see Map Viewer Help, Human Maps Help or Using the Map Viewer to Explore Genomes, a chapter in The NCBI Handbook. Acknowledgments
Continue with other tutorials:
|
||||
Last updated: January 2, 2003
1
![]() 2
2
![]() 3
3
![]() 4
4
![]() 5
5
![]() 6
6
![]() 7
7
![]() 8
8
![]() 9
9
![]() 10
10
![]() 11
11
![]() 12
12
![]() 13
13
![]() 14
14
![]() 15
15
![]() 16
16
![]() 17
17
![]() 18
18
![]() 19
19
![]() 20
20
![]() 21
21
![]() 22
22
![]() X
X ![]() Y
Y
Home ![]() Site
Index
Site
Index ![]() Chromosome
Viewer
Chromosome
Viewer ![]() Genetic
Disorder Guide
Genetic
Disorder Guide ![]() Gene
and Protein Guide
Gene
and Protein Guide ![]() Bioinformatics
Tutorials
Bioinformatics
Tutorials
![]() Bioinformatics
Terms
Bioinformatics
Terms ![]() Sample
Profiles
Sample
Profiles ![]() Evaluating
Medical Information
Evaluating
Medical Information ![]() Links
Links
![]() FAQs
FAQs ![]() Order Poster
Order Poster
 |
The online presentation of this poster is a special feature of the U.S. Department of Energy (DOE) Human Genome Project Information Web site. The DOE Biological and Environmental Research program of the Office of Science funds this site. |