 |
|
Mechanisms of DNA Synthesis Samuel H. Wilson, M.D.
Principal Investigator Tel (919) 541-3267 Fax (919) 541-3592 wilson5@niehs.nih.gov Curriculum Vitae (http://www.niehs.nih.gov/research/atniehs/labs/lsb/dnarna/docs/science-wilson-cv.pdf) Mail Drop B2-06 Research Triangle Park, North Carolina 27709 Delivery Instructions Research SummaryThe DNA Repair & Nucleic Acid Enzymology Group has made a number of contributions over the years to the understanding of mechanisms of DNA synthesis and, in particular, synthesis by the repair enzyme DNA polymerase β (Pol β). They identified the kinetic mechanism of this enzyme, its primary and 3 dimensional structures, and its biological roles in base excision DNA repair. This DNA repair pathway protects cells against endogenous DNA lesion including single-strand breaks. 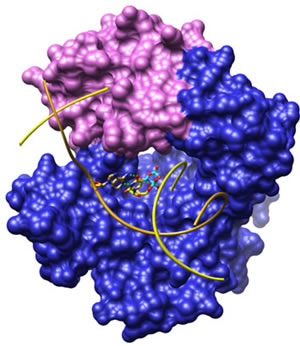 Figure 1. A surface representation of human DNA polymerase β (Pol β), a central enzyme in the base excision repair (BER) pathway
In 1996, the group demonstrated that Pol β "knock-out" mouse fibroblast cell lines are deficient in DNA repair and in cellular protection against DNA damaging agents, thus establishing a cellular role for this enzyme. In 1985, the group discovered the amino acid sequences of human and rat Pol β through cDNA cloning; this was the first such cloning success for any of the mammalian DNA polymerases. The group developed a strong recombinant expression system for the enzyme and then went on to obtain the first crystal structure of a DNA polymerase ternary complex: Pol β complexed with its substrates, DNA and correct incoming dNTP-Mg2+. The group proposed an "induced-fit mechanism" for explaining how DNA polymerases recognized a correct incoming dNTP. Their hypothesis emphasizes conformational changes when a correct incoming nucleotide is positioned in the polymerase active site opposite the templating base. These conformational changes trigger chemistry; the group demonstrated that alteration of key amino acid side chains cripples Pol β so that it no longer catalyzes DNA synthesis efficiently and does not discriminate between correct and incorrect incoming nucleotides. The group is now studying the mechanism of incorrect nucleotides by the enzyme. The group conducts physical, biochemical, cell and molecular biology research on mammalian base excision repair enzymes—in particular, the study of gap-filling DNA synthesis in DNA repair. To define the biological role of DNA polymerase beta, the group and collaborators constructed DNA polymerase "knock-out" cell lines from a transgenic mouse model. These cell lines are devoid of all DNA polymerase β mRNA and protein, and cell extracts lack full base excision repair capacity, thus establishing a role of this particular DNA polymerase in base excision repair. The group and collaborators have reported many crystal structures of complexes of rat and human DNA polymerases beta with the two substrates (DNA and dNTP), and have solved the NMR structure of the enzyme’s 8 kDa domain that contributes lyase activity in the base excision repair pathway. This work has improved understanding of the fundamental mechanism of DNA synthesis and of the phenomenon of templating. The group also studies cellular signaling of DNA damage, as the base excision repair machinery interfaces with cell cycle and cell death signaling. Finally, the research program also includes studies of structure-function relationships of the HIV-1 Reverse Transcriptase. The group and collaborators have conducted extensive kinetic studies of enzyme-nucleic acid interactions. The information from this work provides a framework for drug design and for biochemical analysis of the relationship between the structure of the reverse transcriptase and its functions. Major areas of research:
Current projects:
Collaborators:
Samuel H. Wilson, M.D., heads the DNA Repair & Nucleic Acid Enzymology Group within the Laboratory of Structural Biology. He is also the Deputy Director of the NIEHS and the National Toxicology Program. He received his graduate and postgraduate training in medicine and biochemistry at Harvard Medical School and the NIH. He began his career as a Principal Investigator in 1970 at the NIH. In 1991, Wilson moved to the extramural community to found a center focused in the area of genetic toxicology and functional genomics. He has served in his present positions at NIEHS since the mid-1990s. Wilson has authored and co-authored more than 300 research publications, and has been editor of four reference volumes. |
|

