 |
Bioinformatics: Examining Variation
To view the PDF on this page, you will need Adobe Reader. 
The BLAST program compares a single input sequence, one at
a time, to others in a sequence database. The results can provide
clues as to the identity and function of the input sequence.
Sometimes you may want to compare a number of different
sequences, all at the same time to see where they are alike and
where they are different. The CLUSTAL program was developed
to produce such multiple alignments. CLUSTAL gets its name
because it deals with clusters of sequences.
CLUSTAL alignments are sometimes used by scientists examining
genetic variation within a population. For example, once a gene
has been associated with a disease, scientists can use CLUSTAL
to examine how the gene sequence varies among people with
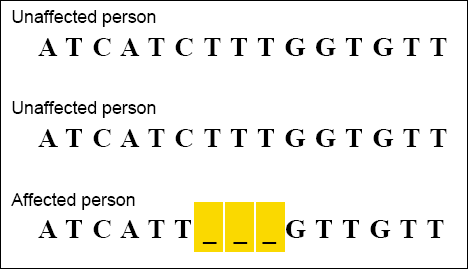
and without the disease. The example below shows a CLUSTAL
alignment of DNA sequences from a portion of the gene associated
with cystic fibrosis. The person affected by the disease is seen to
be missing a three-base DNA sequence.
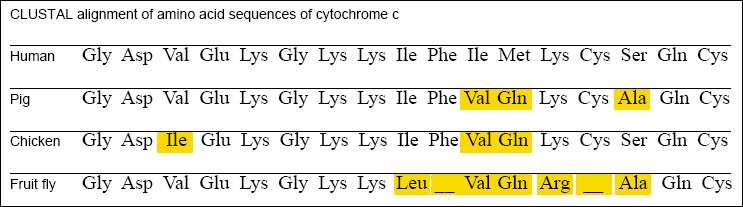
Multiple sequence alignments are also useful to scientists investigating the evolutionary relationships among species. For example, the
CLUSTAL program can be used to align a series of related sequences from different species. Once the program has produced the best
alignment for the sequences, another program can calculate the evolutionary relationships between them. These data can be used to construct a tree diagram showing the evolutionary relationships for that sequence among the various species.
Check Your Understanding
This Bioinformatics lesson contains interactive Check Your Understanding exercises. The exercises are included in the Bioinformatics multimedia download. To complete the exercises: go to the Multimedia Downloads page and download Bioinformatics to your computer.
Last Updated: March 11, 2008
|
 |

|
 |

